Gene
KWMTBOMO08209
Annotation
PREDICTED:_uncharacterized_protein_LOC105841331_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.807
Sequence
CDS
ATGAAGACAGTCAAGCGTCTCGCCCGCGATGAAACTGATTTCCCGATCGCTTCAGAAATCACTAGTGATCATCTTTATATGGATGATCTAGGAGTTTCGGTAGATACGCCCGAGAATGGAATTCAGTTGGCTCATGATTTAATCGAACTATTCAAACGTGGTGGTTTCGACTTAGTTAAATTCGTTAGCAACTCGCCCGAGGTGTTAAAATCGATTCCGCAAGCCAACCGCTTATCTTCAAAAATTGAATTCGACAAGCAGGATAATATGAAGCTCCTAGGTCTAATCTGGTCACCGGTCGAGGATGAATTTTCAATCGCTATTAAATTCAACCCGCAAGCAAAATGTACAAAGCGCCTTATGCTTTCCACAATCGCTAAACTGTGGGACATAAACGGATTTTGTGCCCCTGTCATTTTACACGCAAAATTGTTAATAAAAGCTCTCTGGGTAGCTCAGATTGATTGGGATGCAGAGCCCCCAAGCGACATCTGTGCTGCATGGAAACGCTTTCTTAGTGAACTCCCCGCTCTGCAAGACTTTAAAATACCTCGCTTCGTAGGTATCGTGCCCGACTCCCGTGTCATACTACTGGGATTTGCGGACGCTTCTGAAAAAGCGATGGGTGCTTCTGTGTACATTTGTGTACAACCGGCTTCCGGACCAAATGAAGTCAATTTAATTGGTGCCAAAAGCAAGGTCGCGCCGCTCAAGATTGTTTCCTTAGCCCGCCTAGAGCTTTGTGCTGCTGTATTATTATCGAAACTGTTTCGATTTGTTATCGACACTGTTTCTCCCCGCTGTCATGTTGACGAAATTTTCGCGTTTACTGATTCGACAGTGACCCTCCATTGGGTTCACTCGTCTCCGCATCGCTTTGATACTTTCATTGCGAACCGCATCGCACAAATACAGAACAACCTTGACCCCAAACACTTCTACCATTGTGAAGGTACGCAGAATCCCAGCGACTGCTTGAGTCGTGGTCTCACACCCGCTCAATTATTGAATCACCCGCTGTGGCTCCATGGCCCCACATGGGCTCGTGATGACGTCCATACATGGCCCATCAACCGCTTCACGTTTGATCCCGCACAAGATGAAAAGATATTAGAGCTCAAAAAACAATCAAATGTCTTCATCACTACAGATGAGACAGAACCCTTTCTCTGTACGCTGGCCAAGCGCGTCTCTTCTTGAAAACTCACCGCCTTTACAATGGCCAATGGGCATAATTGA
Protein
MKTVKRLARDETDFPIASEITSDHLYMDDLGVSVDTPENGIQLAHDLIELFKRGGFDLVKFVSNSPEVLKSIPQANRLSSKIEFDKQDNMKLLGLIWSPVEDEFSIAIKFNPQAKCTKRLMLSTIAKLWDINGFCAPVILHAKLLIKALWVAQIDWDAEPPSDICAAWKRFLSELPALQDFKIPRFVGIVPDSRVILLGFADASEKAMGASVYICVQPASGPNEVNLIGAKSKVAPLKIVSLARLELCAAVLLSKLFRFVIDTVSPRCHVDEIFAFTDSTVTLHWVHSSPHRFDTFIANRIAQIQNNLDPKHFYHCEGTQNPSDCLSRGLTPAQLLNHPLWLHGPTWARDDVHTWPINRFTFDPAQDEKILELKKQSNVFITTDETEPFLCTLAKRVSSXKLTAFTMANGHN
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
S4NIN9
A0A1S3DBQ3
A0A1S3DMK8
A0A1S3DTS8
A0A1S4EIX5
A0A1S3DIZ2
+ More
A0A1Y1LIC8 A0A1S3DJJ0 A0A1S3DJ34 A0A1S3DJC0 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A0A9VUS3 A0A0A9X9A3 A0A1S3DM22 A0A1S3DMQ9 A0A1S3DMT1 A0A3Q0JHG9 A0A3Q0JFQ3 V5GHM7 A0A3Q0JJJ7 A0A3Q0J7N1 A0A3Q0J7K9 A0A0A9XZV3 A0A182I0G9 A0A146M0N4 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 A0A1S3DJC4 A0A1B6KIX6 A0A3Q0J0P0 A0A1S3DHM0 A0A3Q0JCL2 A0A1B6MUZ9 A0A1S3DH93 A0A1B6MFR0 A0A1B6KZV2 A0A437AV42 J9JUM9 D7EKN2 A0A146LBU6 A0A0A9W8L8 A0A0J7KGQ6 A0A1Y1KMX0 A0A0J7K7U0 V5GC07 A0A1Y1LM33 A0A1B6GGX5 D7EKN1 A0A1S3DNE5 A0A1Y1LIG5 A0A1B6GMH9 A0A0J7N5Z0 A0A182X947 A0A0J7KNK6 A0A0J7KCI2 D6WY83 A0A3Q0JGS5 A0A0J7JWZ4 J9JNP4 W8BU98 X1XU26 A0A0J7KCV7 V5GY50 A0A0J7K9I5 A0A1B6LDH0 A0A0J7K952 A0A1S3DBK1 A0A0V0Z0L1 A0A2S2PED9 X1WSS7 A0A1S3DQE7 A0A1S3DCY4 A0A146KZL4 A0A0C9PIT9 W8AH72 A0A1B0D8P4 A0A0J7N189 A0A1S3DHK4 X1WPD9 X1WRA3 A0A0A9YD88 A0A224XGH3 A0A0J7N2B8 A0A0J7N465 A0A0J7K7L1 J9KD60 J9JVD6 A0A2S2NNC8 A0A0J7KCR4 A0A0J7KD57 A0A0J7KC70 A0A0A9YQQ8 X1WZY4 A0A1S3DMW8 A0A2S2QPS5 A0A0J7KB34
A0A1Y1LIC8 A0A1S3DJJ0 A0A1S3DJ34 A0A1S3DJC0 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A0A9VUS3 A0A0A9X9A3 A0A1S3DM22 A0A1S3DMQ9 A0A1S3DMT1 A0A3Q0JHG9 A0A3Q0JFQ3 V5GHM7 A0A3Q0JJJ7 A0A3Q0J7N1 A0A3Q0J7K9 A0A0A9XZV3 A0A182I0G9 A0A146M0N4 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 A0A1S3DJC4 A0A1B6KIX6 A0A3Q0J0P0 A0A1S3DHM0 A0A3Q0JCL2 A0A1B6MUZ9 A0A1S3DH93 A0A1B6MFR0 A0A1B6KZV2 A0A437AV42 J9JUM9 D7EKN2 A0A146LBU6 A0A0A9W8L8 A0A0J7KGQ6 A0A1Y1KMX0 A0A0J7K7U0 V5GC07 A0A1Y1LM33 A0A1B6GGX5 D7EKN1 A0A1S3DNE5 A0A1Y1LIG5 A0A1B6GMH9 A0A0J7N5Z0 A0A182X947 A0A0J7KNK6 A0A0J7KCI2 D6WY83 A0A3Q0JGS5 A0A0J7JWZ4 J9JNP4 W8BU98 X1XU26 A0A0J7KCV7 V5GY50 A0A0J7K9I5 A0A1B6LDH0 A0A0J7K952 A0A1S3DBK1 A0A0V0Z0L1 A0A2S2PED9 X1WSS7 A0A1S3DQE7 A0A1S3DCY4 A0A146KZL4 A0A0C9PIT9 W8AH72 A0A1B0D8P4 A0A0J7N189 A0A1S3DHK4 X1WPD9 X1WRA3 A0A0A9YD88 A0A224XGH3 A0A0J7N2B8 A0A0J7N465 A0A0J7K7L1 J9KD60 J9JVD6 A0A2S2NNC8 A0A0J7KCR4 A0A0J7KD57 A0A0J7KC70 A0A0A9YQQ8 X1WZY4 A0A1S3DMW8 A0A2S2QPS5 A0A0J7KB34
EMBL
GAIX01013989
JAA78571.1
GEZM01054789
JAV73399.1
GBBI01004276
JAC14436.1
+ More
GBHO01044644 JAF98959.1 GBHO01044645 JAF98958.1 GBHO01044643 JAF98960.1 GBHO01027393 JAG16211.1 GALX01004947 JAB63519.1 GBHO01020799 JAG22805.1 APCN01002077 GDHC01006469 JAQ12160.1 GEBQ01015458 JAT24519.1 GEBQ01000825 JAT39152.1 GEBQ01028569 JAT11408.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GEBQ01005223 JAT34754.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 RSAL01000392 RVE41994.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 DS497760 EFA13225.1 GDHC01014057 JAQ04572.1 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 LBMM01007770 KMQ89434.1 GEZM01085145 GEZM01085143 JAV60187.1 LBMM01012238 KMQ86334.1 GALX01000776 JAB67690.1 GEZM01054766 GEZM01054765 JAV73410.1 GECZ01008071 JAS61698.1 EFA13221.1 JAV73413.1 GECZ01006169 JAS63600.1 LBMM01009496 KMQ88090.1 LBMM01005079 KMQ91831.1 LBMM01009688 KMQ87939.1 KQ971357 EFA07702.1 LBMM01024307 KMQ82592.1 ABLF02005684 GAMC01009674 JAB96881.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 LBMM01009403 KMQ88162.1 GALX01003138 JAB65328.1 LBMM01011244 KMQ86944.1 GEBQ01018232 JAT21745.1 LBMM01011245 KMQ86943.1 JYDQ01000959 KRY06068.1 GGMR01015135 MBY27754.1 ABLF02017056 GDHC01018192 JAQ00437.1 GBYB01000873 JAG70640.1 GAMC01021577 GAMC01021573 JAB84982.1 AJVK01027627 LBMM01012077 KMQ86420.1 ABLF02006985 ABLF02066034 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 GFTR01008866 JAW07560.1 LBMM01011492 KMQ86800.1 LBMM01010417 KMQ87475.1 LBMM01012188 KMQ86362.1 ABLF02041863 ABLF02008594 GGMR01005823 MBY18442.1 LBMM01009551 KMQ88039.1 LBMM01009442 KMQ88136.1 LBMM01009863 KMQ87831.1 GBHO01009080 JAG34524.1 ABLF02036131 ABLF02041527 ABLF02064049 GGMS01010524 MBY79727.1 LBMM01010405 KMQ87487.1
GBHO01044644 JAF98959.1 GBHO01044645 JAF98958.1 GBHO01044643 JAF98960.1 GBHO01027393 JAG16211.1 GALX01004947 JAB63519.1 GBHO01020799 JAG22805.1 APCN01002077 GDHC01006469 JAQ12160.1 GEBQ01015458 JAT24519.1 GEBQ01000825 JAT39152.1 GEBQ01028569 JAT11408.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GEBQ01005223 JAT34754.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 RSAL01000392 RVE41994.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 DS497760 EFA13225.1 GDHC01014057 JAQ04572.1 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 LBMM01007770 KMQ89434.1 GEZM01085145 GEZM01085143 JAV60187.1 LBMM01012238 KMQ86334.1 GALX01000776 JAB67690.1 GEZM01054766 GEZM01054765 JAV73410.1 GECZ01008071 JAS61698.1 EFA13221.1 JAV73413.1 GECZ01006169 JAS63600.1 LBMM01009496 KMQ88090.1 LBMM01005079 KMQ91831.1 LBMM01009688 KMQ87939.1 KQ971357 EFA07702.1 LBMM01024307 KMQ82592.1 ABLF02005684 GAMC01009674 JAB96881.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 LBMM01009403 KMQ88162.1 GALX01003138 JAB65328.1 LBMM01011244 KMQ86944.1 GEBQ01018232 JAT21745.1 LBMM01011245 KMQ86943.1 JYDQ01000959 KRY06068.1 GGMR01015135 MBY27754.1 ABLF02017056 GDHC01018192 JAQ00437.1 GBYB01000873 JAG70640.1 GAMC01021577 GAMC01021573 JAB84982.1 AJVK01027627 LBMM01012077 KMQ86420.1 ABLF02006985 ABLF02066034 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 GFTR01008866 JAW07560.1 LBMM01011492 KMQ86800.1 LBMM01010417 KMQ87475.1 LBMM01012188 KMQ86362.1 ABLF02041863 ABLF02008594 GGMR01005823 MBY18442.1 LBMM01009551 KMQ88039.1 LBMM01009442 KMQ88136.1 LBMM01009863 KMQ87831.1 GBHO01009080 JAG34524.1 ABLF02036131 ABLF02041527 ABLF02064049 GGMS01010524 MBY79727.1 LBMM01010405 KMQ87487.1
Proteomes
Pfam
Interpro
IPR041588
Integrase_H2C2
+ More
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001584 Integrase_cat-core
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR008737 Peptidase_asp_put
IPR000477 RT_dom
IPR005312 DUF1759
IPR021109 Peptidase_aspartic_dom_sf
IPR011012 Longin-like_dom_sf
IPR010908 Longin_dom
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR032403 Exo84_C
IPR016159 Cullin_repeat-like_dom_sf
IPR000727 T_SNARE_dom
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR001969 Aspartic_peptidase_AS
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001584 Integrase_cat-core
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR008737 Peptidase_asp_put
IPR000477 RT_dom
IPR005312 DUF1759
IPR021109 Peptidase_aspartic_dom_sf
IPR011012 Longin-like_dom_sf
IPR010908 Longin_dom
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR032403 Exo84_C
IPR016159 Cullin_repeat-like_dom_sf
IPR000727 T_SNARE_dom
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR001969 Aspartic_peptidase_AS
SUPFAM
Gene 3D
ProteinModelPortal
S4NIN9
A0A1S3DBQ3
A0A1S3DMK8
A0A1S3DTS8
A0A1S4EIX5
A0A1S3DIZ2
+ More
A0A1Y1LIC8 A0A1S3DJJ0 A0A1S3DJ34 A0A1S3DJC0 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A0A9VUS3 A0A0A9X9A3 A0A1S3DM22 A0A1S3DMQ9 A0A1S3DMT1 A0A3Q0JHG9 A0A3Q0JFQ3 V5GHM7 A0A3Q0JJJ7 A0A3Q0J7N1 A0A3Q0J7K9 A0A0A9XZV3 A0A182I0G9 A0A146M0N4 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 A0A1S3DJC4 A0A1B6KIX6 A0A3Q0J0P0 A0A1S3DHM0 A0A3Q0JCL2 A0A1B6MUZ9 A0A1S3DH93 A0A1B6MFR0 A0A1B6KZV2 A0A437AV42 J9JUM9 D7EKN2 A0A146LBU6 A0A0A9W8L8 A0A0J7KGQ6 A0A1Y1KMX0 A0A0J7K7U0 V5GC07 A0A1Y1LM33 A0A1B6GGX5 D7EKN1 A0A1S3DNE5 A0A1Y1LIG5 A0A1B6GMH9 A0A0J7N5Z0 A0A182X947 A0A0J7KNK6 A0A0J7KCI2 D6WY83 A0A3Q0JGS5 A0A0J7JWZ4 J9JNP4 W8BU98 X1XU26 A0A0J7KCV7 V5GY50 A0A0J7K9I5 A0A1B6LDH0 A0A0J7K952 A0A1S3DBK1 A0A0V0Z0L1 A0A2S2PED9 X1WSS7 A0A1S3DQE7 A0A1S3DCY4 A0A146KZL4 A0A0C9PIT9 W8AH72 A0A1B0D8P4 A0A0J7N189 A0A1S3DHK4 X1WPD9 X1WRA3 A0A0A9YD88 A0A224XGH3 A0A0J7N2B8 A0A0J7N465 A0A0J7K7L1 J9KD60 J9JVD6 A0A2S2NNC8 A0A0J7KCR4 A0A0J7KD57 A0A0J7KC70 A0A0A9YQQ8 X1WZY4 A0A1S3DMW8 A0A2S2QPS5 A0A0J7KB34
A0A1Y1LIC8 A0A1S3DJJ0 A0A1S3DJ34 A0A1S3DJC0 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A0A9VUS3 A0A0A9X9A3 A0A1S3DM22 A0A1S3DMQ9 A0A1S3DMT1 A0A3Q0JHG9 A0A3Q0JFQ3 V5GHM7 A0A3Q0JJJ7 A0A3Q0J7N1 A0A3Q0J7K9 A0A0A9XZV3 A0A182I0G9 A0A146M0N4 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 A0A1S3DJC4 A0A1B6KIX6 A0A3Q0J0P0 A0A1S3DHM0 A0A3Q0JCL2 A0A1B6MUZ9 A0A1S3DH93 A0A1B6MFR0 A0A1B6KZV2 A0A437AV42 J9JUM9 D7EKN2 A0A146LBU6 A0A0A9W8L8 A0A0J7KGQ6 A0A1Y1KMX0 A0A0J7K7U0 V5GC07 A0A1Y1LM33 A0A1B6GGX5 D7EKN1 A0A1S3DNE5 A0A1Y1LIG5 A0A1B6GMH9 A0A0J7N5Z0 A0A182X947 A0A0J7KNK6 A0A0J7KCI2 D6WY83 A0A3Q0JGS5 A0A0J7JWZ4 J9JNP4 W8BU98 X1XU26 A0A0J7KCV7 V5GY50 A0A0J7K9I5 A0A1B6LDH0 A0A0J7K952 A0A1S3DBK1 A0A0V0Z0L1 A0A2S2PED9 X1WSS7 A0A1S3DQE7 A0A1S3DCY4 A0A146KZL4 A0A0C9PIT9 W8AH72 A0A1B0D8P4 A0A0J7N189 A0A1S3DHK4 X1WPD9 X1WRA3 A0A0A9YD88 A0A224XGH3 A0A0J7N2B8 A0A0J7N465 A0A0J7K7L1 J9KD60 J9JVD6 A0A2S2NNC8 A0A0J7KCR4 A0A0J7KD57 A0A0J7KC70 A0A0A9YQQ8 X1WZY4 A0A1S3DMW8 A0A2S2QPS5 A0A0J7KB34
Ontologies
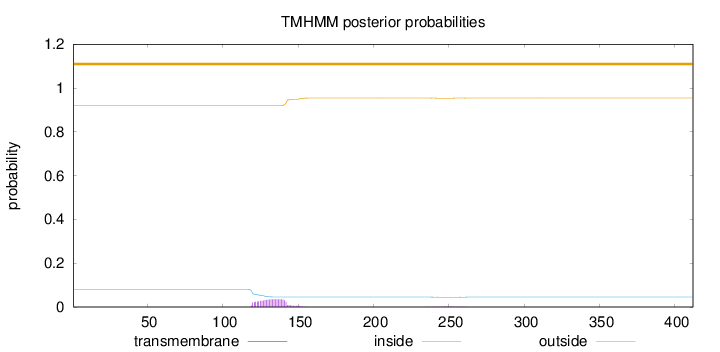
Topology
Length:
412
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.7992
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08065
outside
1 - 412
Population Genetic Test Statistics
Pi
300.114394
Theta
145.341425
Tajima's D
3.658906
CLR
6.866997
CSRT
0.995650217489126
Interpretation
Uncertain
