Gene
KWMTBOMO08205
Pre Gene Modal
BGIBMGA009465
Annotation
PREDICTED:_tRNA:m(4)X_modification_enzyme_TRM13_homolog_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.245 Mitochondrial Reliability : 1.155 Nuclear Reliability : 1.739
Sequence
CDS
ATGTCCAAAAATATACACTCTACTAAGGAATCAAACGGACCTCAATGCCAGTACTTTGTACTGAGGAAAAAACGGCTGTGCCGTATGACTGTGCGTCCTGGCAATCAGTACTGCGGTGAACATCAACCCCCACCTAATGATTGCTTGTCTGACAAACGTATTGCATGTCCCAACGATCCTAAGCACACATGCTACATTAGCAAATTGGAAAAGCATCTAACCATCTGTAATGCTAGACAACAAGACCAGCCGCCCTACATTGAATACAACATCAATACTCCGATAACAAATGCAGTTCCTCGAATTTTGCTCAGCGAGATCCCTGGTGATATAATCATTCAGGTCATAAATAAAGTTAATTCATTATATGACAAATTTATCAAATCGAACATAGTATCAATACCAGAGCGCGCTATCCATGCTGCTGTAGCACCAGAGTTCAATGAGGCGGATCGGCAGGAGAGCTCCCGCCGACATTTGCGGCAAACATCTAGCTTGTTATGGCTCGTGGAAGAAGAGAACCTAGTGGATAATGAGACTTGCTATGTGGAACTTGGAGCTGGCAAAGGGCAGCTGTCGTTCTACGCGCACACGGCGTGGTGCGCGGGCGCGGGGTCCAGCGTGCTGCTGGTGGACCGCGCCACGCTGCGGCACAAGCGCGACGGGCGGCTGCGGGGGCGGGGCGCCGCGCGGCTGCGGGCCGAGTTGGCGCACCTGGCGCTGCACCGCGTGCCGGCCGCCGCCACCGCGCGCCGCCTCGTGGGCCTGGCCAAGCACCTGTGCGGCGTGGCCACCGACTACGCGCTGCGGTGCATGACATCGGAGGGCGTGCTGGGGAAGACCAGAGGCGTGGTGATCGCGACTTGCTGCCACCACCGCTGCGAATACTCCGCTTATGTCGGCAACCGACACCTACTGGACATGGGTGTAACCGCCGAAGACTTGAACGCGATGCTCGGGCTGGTGTCCTGGGCCACGTGCGGGGACGGCCGCAACCGACTCCACCGACAGAACAATCCGGCCAGCGACGAGCACTTAGCGACGACCGAAACCGATCACTCCAACAGTGAAGTCGGTTGTGACAATAATGTACCGGACGAGGCGCCCCCCGCCGCCGCAACCCACGAACAGCTGAAGCACAGCCTCAGTCGCGAGCAGAGGGAGCAGGTCGGCAGGAGAGCTAAGGCGCTGCTGGACTGGGGCAGGAAACTCTACTTAGAAGAAAAGGGCTTCAACGTGAAACTGTGTCACTACGTGCCCATCGGAGTGTCGCCGGAGAACGTCTGTATAGTCGCCACGAAAATGTGA
Protein
MSKNIHSTKESNGPQCQYFVLRKKRLCRMTVRPGNQYCGEHQPPPNDCLSDKRIACPNDPKHTCYISKLEKHLTICNARQQDQPPYIEYNINTPITNAVPRILLSEIPGDIIIQVINKVNSLYDKFIKSNIVSIPERAIHAAVAPEFNEADRQESSRRHLRQTSSLLWLVEEENLVDNETCYVELGAGKGQLSFYAHTAWCAGAGSSVLLVDRATLRHKRDGRLRGRGAARLRAELAHLALHRVPAAATARRLVGLAKHLCGVATDYALRCMTSEGVLGKTRGVVIATCCHHRCEYSAYVGNRHLLDMGVTAEDLNAMLGLVSWATCGDGRNRLHRQNNPASDEHLATTETDHSNSEVGCDNNVPDEAPPAAATHEQLKHSLSREQREQVGRRAKALLDWGRKLYLEEKGFNVKLCHYVPIGVSPENVCIVATKM
Summary
Uniprot
H9JIW4
A0A2H1WUJ5
A0A212EU66
D7EIE5
A0A1Y1K2W0
A0A194PZ51
+ More
A0A026W2Y0 A0A158NEA2 E2A0A8 A0A151WSL1 Q16QR4 B0WSZ0 E2B648 U4UNG7 A0A0N8P1N6 A0A195FM28 A0A0L7R8E7 A0A151J0H1 A0A1W4V583 B3LVS8 A0A310SCU3 A0A1W4WH75 A0A1W4WS31 A0A182GM31 A0A0J7NHK8 F4WJC4 A0A0N0BBH0 A0A2J7RMD2 A0A088A5C4 A0A034VND0 A0A182XY74 A0A084VVB8 A0A0K8UED4 A0A0K8UKQ8 A0A1Q3F5D4 A0A182I9W5 A0A182SHS3 A0A154P931 A0A182TI60 B3P2K2 W8C6V1 Q7QEF3 A0A182VGU3 A0A0L0BQE4 A0A182X7H5 B4G3C1 A0A1W4WT32 A0A1I8NQ20 A0A182QDB4 B4MBN2 A0A182MZZ8 Q8SX24 B4KBP2 Q298B0 B4PVQ2 A0A1B0CVW7 A0A1B0FL81 A0A3B0JHK3 B4NHB3 A0A0A1XKJ1 K7J650 A0A2A3E537 A0A1B0B2Z3 B4I4E9 A0A0M4EQX1 A0A1A9VDP5 A0A1A9W5N8 A0A1I8MJT4 A0A232EVD5 A0A182J888 A0A182RYS4 A0A1A9ZSV2 A0A2M4ALY9 A0A1B0D3U1 A0A2M4CR58 A0A2M4CR64 A0A2M4CRB1 U5EU63 B4JYD5 A0A182MN80 A0A336MJC2 A0A1J1IQV7 A0A1A9YQ65 A0A0P4WC82 B4QYA5 A0A3R7SSN4 A0A0L8H6D4 A0A182FWE6 A0A0P5J474 A0A3M6THD2 A0A0P5FWJ7 A0A0P6GBY3 C3YCH1 A0A2B4S021
A0A026W2Y0 A0A158NEA2 E2A0A8 A0A151WSL1 Q16QR4 B0WSZ0 E2B648 U4UNG7 A0A0N8P1N6 A0A195FM28 A0A0L7R8E7 A0A151J0H1 A0A1W4V583 B3LVS8 A0A310SCU3 A0A1W4WH75 A0A1W4WS31 A0A182GM31 A0A0J7NHK8 F4WJC4 A0A0N0BBH0 A0A2J7RMD2 A0A088A5C4 A0A034VND0 A0A182XY74 A0A084VVB8 A0A0K8UED4 A0A0K8UKQ8 A0A1Q3F5D4 A0A182I9W5 A0A182SHS3 A0A154P931 A0A182TI60 B3P2K2 W8C6V1 Q7QEF3 A0A182VGU3 A0A0L0BQE4 A0A182X7H5 B4G3C1 A0A1W4WT32 A0A1I8NQ20 A0A182QDB4 B4MBN2 A0A182MZZ8 Q8SX24 B4KBP2 Q298B0 B4PVQ2 A0A1B0CVW7 A0A1B0FL81 A0A3B0JHK3 B4NHB3 A0A0A1XKJ1 K7J650 A0A2A3E537 A0A1B0B2Z3 B4I4E9 A0A0M4EQX1 A0A1A9VDP5 A0A1A9W5N8 A0A1I8MJT4 A0A232EVD5 A0A182J888 A0A182RYS4 A0A1A9ZSV2 A0A2M4ALY9 A0A1B0D3U1 A0A2M4CR58 A0A2M4CR64 A0A2M4CRB1 U5EU63 B4JYD5 A0A182MN80 A0A336MJC2 A0A1J1IQV7 A0A1A9YQ65 A0A0P4WC82 B4QYA5 A0A3R7SSN4 A0A0L8H6D4 A0A182FWE6 A0A0P5J474 A0A3M6THD2 A0A0P5FWJ7 A0A0P6GBY3 C3YCH1 A0A2B4S021
Pubmed
19121390
22118469
18362917
19820115
28004739
26354079
+ More
24508170 30249741 21347285 20798317 17510324 23537049 17994087 26483478 21719571 25348373 25244985 24438588 24495485 12364791 14747013 17210077 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 25830018 20075255 25315136 28648823 30382153 18563158
24508170 30249741 21347285 20798317 17510324 23537049 17994087 26483478 21719571 25348373 25244985 24438588 24495485 12364791 14747013 17210077 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 25830018 20075255 25315136 28648823 30382153 18563158
EMBL
BABH01023052
ODYU01011129
SOQ56677.1
AGBW02012474
OWR45001.1
DS497678
+ More
EFA11905.1 GEZM01096538 GEZM01096534 JAV54831.1 KQ459585 KPI98318.1 KK107455 QOIP01000010 EZA50432.1 RLU17668.1 ADTU01013183 ADTU01013184 GL435569 EFN73111.1 KQ982769 KYQ50846.1 CH477736 EAT36739.1 DS232078 EDS34123.1 GL445914 EFN88837.1 KB632375 ERL94013.1 CH902617 KPU80428.1 KQ981490 KYN41039.1 KQ414633 KOC67036.1 KQ980636 KYN14952.1 EDV43702.1 KQ777311 OAD52137.1 JXUM01073756 KQ562794 KXJ75118.1 LBMM01004910 KMQ91995.1 GL888182 EGI65702.1 KQ436019 KOX67568.1 NEVH01002562 PNF41990.1 GAKP01014968 JAC43984.1 ATLV01017170 KE525157 KFB41912.1 GDHF01027383 JAI24931.1 GDHF01025374 JAI26940.1 GFDL01012264 JAV22781.1 APCN01001063 KQ434846 KZC08352.1 CH954181 EDV48024.1 GAMC01004024 JAC02532.1 AAAB01008847 EAA06835.4 JRES01001623 KNC21424.1 CH479179 EDW24303.1 AXCN02001335 CH940656 EDW58503.2 AE014297 AY094893 AAF54134.2 AAM11246.1 CH933806 EDW14719.2 CM000070 EAL28045.1 CM000160 EDW97861.2 AJWK01031360 CCAG010013281 OUUW01000005 SPP80183.1 CH964272 EDW84589.2 GBXI01002825 JAD11467.1 KZ288364 PBC26873.1 JXJN01007738 CH480821 EDW55092.1 CP012526 ALC46810.1 NNAY01002011 OXU22314.1 GGFK01008431 MBW41752.1 AJVK01023693 GGFL01003638 MBW67816.1 GGFL01003639 MBW67817.1 GGFL01003637 MBW67815.1 GANO01001629 JAB58242.1 CH916377 EDV90697.1 AXCM01002614 UFQS01001315 UFQT01001315 SSX10307.1 SSX29995.1 CVRI01000055 CRL01478.1 GDRN01070236 JAI63915.1 CM000364 EDX11862.1 QCYY01002044 ROT73279.1 KQ419049 KOF84772.1 GDIQ01205469 JAK46256.1 RCHS01003600 RMX40654.1 GDIQ01249399 GDIQ01227908 JAK02326.1 GDIQ01208166 GDIQ01035573 JAN59164.1 GG666501 EEN62008.1 LSMT01000230 PFX22756.1
EFA11905.1 GEZM01096538 GEZM01096534 JAV54831.1 KQ459585 KPI98318.1 KK107455 QOIP01000010 EZA50432.1 RLU17668.1 ADTU01013183 ADTU01013184 GL435569 EFN73111.1 KQ982769 KYQ50846.1 CH477736 EAT36739.1 DS232078 EDS34123.1 GL445914 EFN88837.1 KB632375 ERL94013.1 CH902617 KPU80428.1 KQ981490 KYN41039.1 KQ414633 KOC67036.1 KQ980636 KYN14952.1 EDV43702.1 KQ777311 OAD52137.1 JXUM01073756 KQ562794 KXJ75118.1 LBMM01004910 KMQ91995.1 GL888182 EGI65702.1 KQ436019 KOX67568.1 NEVH01002562 PNF41990.1 GAKP01014968 JAC43984.1 ATLV01017170 KE525157 KFB41912.1 GDHF01027383 JAI24931.1 GDHF01025374 JAI26940.1 GFDL01012264 JAV22781.1 APCN01001063 KQ434846 KZC08352.1 CH954181 EDV48024.1 GAMC01004024 JAC02532.1 AAAB01008847 EAA06835.4 JRES01001623 KNC21424.1 CH479179 EDW24303.1 AXCN02001335 CH940656 EDW58503.2 AE014297 AY094893 AAF54134.2 AAM11246.1 CH933806 EDW14719.2 CM000070 EAL28045.1 CM000160 EDW97861.2 AJWK01031360 CCAG010013281 OUUW01000005 SPP80183.1 CH964272 EDW84589.2 GBXI01002825 JAD11467.1 KZ288364 PBC26873.1 JXJN01007738 CH480821 EDW55092.1 CP012526 ALC46810.1 NNAY01002011 OXU22314.1 GGFK01008431 MBW41752.1 AJVK01023693 GGFL01003638 MBW67816.1 GGFL01003639 MBW67817.1 GGFL01003637 MBW67815.1 GANO01001629 JAB58242.1 CH916377 EDV90697.1 AXCM01002614 UFQS01001315 UFQT01001315 SSX10307.1 SSX29995.1 CVRI01000055 CRL01478.1 GDRN01070236 JAI63915.1 CM000364 EDX11862.1 QCYY01002044 ROT73279.1 KQ419049 KOF84772.1 GDIQ01205469 JAK46256.1 RCHS01003600 RMX40654.1 GDIQ01249399 GDIQ01227908 JAK02326.1 GDIQ01208166 GDIQ01035573 JAN59164.1 GG666501 EEN62008.1 LSMT01000230 PFX22756.1
Proteomes
UP000005204
UP000007151
UP000007266
UP000053268
UP000053097
UP000279307
+ More
UP000005205 UP000000311 UP000075809 UP000008820 UP000002320 UP000008237 UP000030742 UP000007801 UP000078541 UP000053825 UP000078492 UP000192221 UP000192223 UP000069940 UP000249989 UP000036403 UP000007755 UP000053105 UP000235965 UP000005203 UP000076408 UP000030765 UP000075840 UP000075901 UP000076502 UP000075902 UP000008711 UP000007062 UP000075903 UP000037069 UP000076407 UP000008744 UP000095300 UP000075886 UP000008792 UP000075884 UP000000803 UP000009192 UP000001819 UP000002282 UP000092461 UP000092444 UP000268350 UP000007798 UP000002358 UP000242457 UP000092460 UP000001292 UP000092553 UP000078200 UP000091820 UP000095301 UP000215335 UP000075880 UP000075900 UP000092445 UP000092462 UP000001070 UP000075883 UP000183832 UP000092443 UP000000304 UP000283509 UP000053454 UP000069272 UP000275408 UP000001554 UP000225706
UP000005205 UP000000311 UP000075809 UP000008820 UP000002320 UP000008237 UP000030742 UP000007801 UP000078541 UP000053825 UP000078492 UP000192221 UP000192223 UP000069940 UP000249989 UP000036403 UP000007755 UP000053105 UP000235965 UP000005203 UP000076408 UP000030765 UP000075840 UP000075901 UP000076502 UP000075902 UP000008711 UP000007062 UP000075903 UP000037069 UP000076407 UP000008744 UP000095300 UP000075886 UP000008792 UP000075884 UP000000803 UP000009192 UP000001819 UP000002282 UP000092461 UP000092444 UP000268350 UP000007798 UP000002358 UP000242457 UP000092460 UP000001292 UP000092553 UP000078200 UP000091820 UP000095301 UP000215335 UP000075880 UP000075900 UP000092445 UP000092462 UP000001070 UP000075883 UP000183832 UP000092443 UP000000304 UP000283509 UP000053454 UP000069272 UP000275408 UP000001554 UP000225706
Pfam
Interpro
IPR022776
TRM13/UPF0224_CHHC_Znf_dom
+ More
IPR007871 Methyltransferase_TRM13
IPR021721 Znf_CCCH-type_TRM13
IPR039044 Trm13
IPR029063 SAM-dependent_MTases
IPR000727 T_SNARE_dom
IPR011650 Peptidase_M20_dimer
IPR036264 Bact_exopeptidase_dim_dom
IPR002933 Peptidase_M20
IPR010159 N-acyl_aa_amidohydrolase
IPR007871 Methyltransferase_TRM13
IPR021721 Znf_CCCH-type_TRM13
IPR039044 Trm13
IPR029063 SAM-dependent_MTases
IPR000727 T_SNARE_dom
IPR011650 Peptidase_M20_dimer
IPR036264 Bact_exopeptidase_dim_dom
IPR002933 Peptidase_M20
IPR010159 N-acyl_aa_amidohydrolase
ProteinModelPortal
H9JIW4
A0A2H1WUJ5
A0A212EU66
D7EIE5
A0A1Y1K2W0
A0A194PZ51
+ More
A0A026W2Y0 A0A158NEA2 E2A0A8 A0A151WSL1 Q16QR4 B0WSZ0 E2B648 U4UNG7 A0A0N8P1N6 A0A195FM28 A0A0L7R8E7 A0A151J0H1 A0A1W4V583 B3LVS8 A0A310SCU3 A0A1W4WH75 A0A1W4WS31 A0A182GM31 A0A0J7NHK8 F4WJC4 A0A0N0BBH0 A0A2J7RMD2 A0A088A5C4 A0A034VND0 A0A182XY74 A0A084VVB8 A0A0K8UED4 A0A0K8UKQ8 A0A1Q3F5D4 A0A182I9W5 A0A182SHS3 A0A154P931 A0A182TI60 B3P2K2 W8C6V1 Q7QEF3 A0A182VGU3 A0A0L0BQE4 A0A182X7H5 B4G3C1 A0A1W4WT32 A0A1I8NQ20 A0A182QDB4 B4MBN2 A0A182MZZ8 Q8SX24 B4KBP2 Q298B0 B4PVQ2 A0A1B0CVW7 A0A1B0FL81 A0A3B0JHK3 B4NHB3 A0A0A1XKJ1 K7J650 A0A2A3E537 A0A1B0B2Z3 B4I4E9 A0A0M4EQX1 A0A1A9VDP5 A0A1A9W5N8 A0A1I8MJT4 A0A232EVD5 A0A182J888 A0A182RYS4 A0A1A9ZSV2 A0A2M4ALY9 A0A1B0D3U1 A0A2M4CR58 A0A2M4CR64 A0A2M4CRB1 U5EU63 B4JYD5 A0A182MN80 A0A336MJC2 A0A1J1IQV7 A0A1A9YQ65 A0A0P4WC82 B4QYA5 A0A3R7SSN4 A0A0L8H6D4 A0A182FWE6 A0A0P5J474 A0A3M6THD2 A0A0P5FWJ7 A0A0P6GBY3 C3YCH1 A0A2B4S021
A0A026W2Y0 A0A158NEA2 E2A0A8 A0A151WSL1 Q16QR4 B0WSZ0 E2B648 U4UNG7 A0A0N8P1N6 A0A195FM28 A0A0L7R8E7 A0A151J0H1 A0A1W4V583 B3LVS8 A0A310SCU3 A0A1W4WH75 A0A1W4WS31 A0A182GM31 A0A0J7NHK8 F4WJC4 A0A0N0BBH0 A0A2J7RMD2 A0A088A5C4 A0A034VND0 A0A182XY74 A0A084VVB8 A0A0K8UED4 A0A0K8UKQ8 A0A1Q3F5D4 A0A182I9W5 A0A182SHS3 A0A154P931 A0A182TI60 B3P2K2 W8C6V1 Q7QEF3 A0A182VGU3 A0A0L0BQE4 A0A182X7H5 B4G3C1 A0A1W4WT32 A0A1I8NQ20 A0A182QDB4 B4MBN2 A0A182MZZ8 Q8SX24 B4KBP2 Q298B0 B4PVQ2 A0A1B0CVW7 A0A1B0FL81 A0A3B0JHK3 B4NHB3 A0A0A1XKJ1 K7J650 A0A2A3E537 A0A1B0B2Z3 B4I4E9 A0A0M4EQX1 A0A1A9VDP5 A0A1A9W5N8 A0A1I8MJT4 A0A232EVD5 A0A182J888 A0A182RYS4 A0A1A9ZSV2 A0A2M4ALY9 A0A1B0D3U1 A0A2M4CR58 A0A2M4CR64 A0A2M4CRB1 U5EU63 B4JYD5 A0A182MN80 A0A336MJC2 A0A1J1IQV7 A0A1A9YQ65 A0A0P4WC82 B4QYA5 A0A3R7SSN4 A0A0L8H6D4 A0A182FWE6 A0A0P5J474 A0A3M6THD2 A0A0P5FWJ7 A0A0P6GBY3 C3YCH1 A0A2B4S021
Ontologies
GO
PANTHER
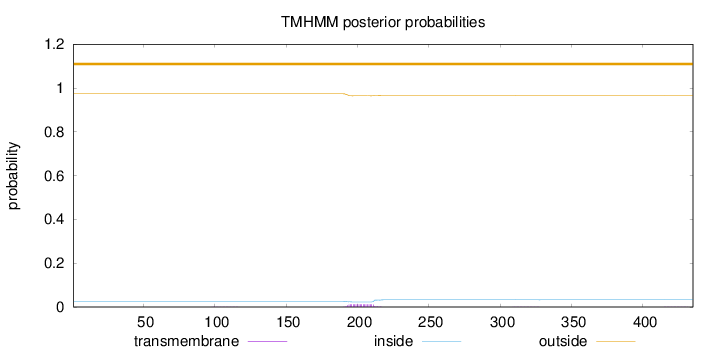
Topology
Length:
435
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25633
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02622
outside
1 - 435
Population Genetic Test Statistics
Pi
200.544848
Theta
158.820064
Tajima's D
1.092396
CLR
0.050379
CSRT
0.691615419229039
Interpretation
Uncertain
