Gene
KWMTBOMO08201 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009467
Annotation
PREDICTED:_glucosamine-6-phosphate_isomerase_isoform_X2_[Papilio_machaon]
Full name
Glucosamine-6-phosphate isomerase
+ More
Glucosamine-6-phosphate isomerase 2
Glucosamine-6-phosphate isomerase 2
Alternative Name
Glucosamine-6-phosphate deaminase
Glucosamine-6-phosphate deaminase 2
Glucosamine-6-phosphate deaminase 2
Location in the cell
Cytoplasmic Reliability : 3.04
Sequence
CDS
ATGCGTCTTATAATTCTCGAAGATGCCAGCATCGTTGCCGACTGGGCCGCCAGATTCGTTCTCCAGCGGATCACGCAGTTCGCTCCGGGTCCGGGAAGACACTTCGTGCTGGGCCTGCCCACCGGTGGGACGCCCCTCGGCATGTACAAACGGTTGATAGAGTTCCACAAGGAAGGGAAGTTATCTTTCAAATATGTGACCACTTTCAACATGGACGAGTATGTGGGTCTTCCCCGTGACCATCCCGAGTCCTACCACTACTACATGTGGAACGAGTTCTTCAAACACATCGACATAGAGCCGAGCAACGCGCACGTGCTCGACGGCAACGCATCCGATTTAGTGCTCGAGTGTCAACGCTTTGAGAAACTCATACAGGAGGCTGGCGGAGTGCATTTGTTCATCGGAGGTATCGGTCCGGACGGTCACATCGCGTTCAACGAACCGGGCTCCTCGCTGGTGTCTCGGACGCGAGTGAAGACCCTGGCCTACGACACCCTGGACGCCAACAAGCGGTTCTTTGACAACGACATCAGCAAGGTGCCCCGACAGGCGCTCACCGTTGGAGTCGGAACTGTTATGGACGCTAAAGAGGTCATGATCCTCATAACAGGAGTGCACAAGTCCTTGGCGCTTGCCAAGGCTGTGGAGGAAGGCGTGAACCACATGTGGACGGTGTCGGCCTTCCAGCAACATCCTCAAGCGCTTTTCGTGTGTGACGAAGACGCCACACTCGAGTTGAGGGTCAAAACAGTTAAATACTTTAAGAGTTTAATGGTGGAACACAATAAACTGATAGAAGCAAAACAATGA
Protein
MRLIILEDASIVADWAARFVLQRITQFAPGPGRHFVLGLPTGGTPLGMYKRLIEFHKEGKLSFKYVTTFNMDEYVGLPRDHPESYHYYMWNEFFKHIDIEPSNAHVLDGNASDLVLECQRFEKLIQEAGGVHLFIGGIGPDGHIAFNEPGSSLVSRTRVKTLAYDTLDANKRFFDNDISKVPRQALTVGVGTVMDAKEVMILITGVHKSLALAKAVEEGVNHMWTVSAFQQHPQALFVCDEDATLELRVKTVKYFKSLMVEHNKLIEAKQ
Summary
Catalytic Activity
alpha-D-glucosamine 6-phosphate + H2O = beta-D-fructose 6-phosphate + NH4(+)
Subunit
Homohexamer.
Similarity
Belongs to the glucosamine/galactosamine-6-phosphate isomerase family.
Keywords
Carbohydrate metabolism
Coiled coil
Complete proteome
Cytoplasm
Hydrolase
Reference proteome
Alternative splicing
Feature
chain Glucosamine-6-phosphate isomerase 2
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9JIW6
A0A222AJ35
A0A2A4JPG6
A0A2H1VU62
A0A437B3V5
A0A2A4JPB1
+ More
A0A212EWA6 A0A2W1BEV1 A0A194PZ46 S4PE45 A0A1J1IPS7 D2A2L2 A0A336LU61 A0A0Q9WBD5 A0A0P8XV28 A0A034WAS0 A0A0K8VDI5 A0A3B0KEY2 A0A0Q9X5S1 A0A1L5L9D6 A0A0R3NXV6 A0A0A1WQ03 A0A1W4XHB0 W8BZB0 A0A0C9RLV6 M9ND31 A0A1B6F5M2 A0A0M4E313 A0A1W4UIY2 V5FZ40 A0A336LLF8 A0A0R1DSK6 A0A1I8MDQ5 A0A034WDD6 A0A0K8UEW5 A0A1B6H960 W8BS96 U5EY06 A0A0Q5WIN9 E0VTH1 A0A0J9TG94 A0A0A1XQ14 A4IHW6 B4LU11 A0A2J7PZD3 A0A0P8ZES1 B4KF38 A0A1L8E1Z9 A0A2P8Y487 A0A0P5S8V7 B3MMZ7 A0A0L0CAZ7 A0A0R3NRD4 A0A1B6DAT5 B4GJK4 A0A3B0K811 A0A1L8HTM5 Q6PA43 E9G0Q9 A0A1B6L607 A0A1I8PZ69 Q29NT9 D3TP29 A0A0P5XS92 A0A1B6KT34 A0A1I8MDR2 A0A1Y1L0K0 B0WKW4 A0A250YE98 A0A411G6A4 W5J8R2 A0A158P2B3 A0A3Q3J3M3 A0A2U3V426 A0A340WTF3 A0A2Y9MIC1 A0A384A0B1 A0A2Y9TFS2 A0A341AA49 Q16HW7-3 F1S3T0 A0A1S2ZYD2 A0A2M4BWA9 A0A2K5RPG0 A0A195D4E5 G3VGH7 A0A1B0D5D0 F4W6K8 A0A2M4ALC9 E2RH59 A0A3Q7UXB9 H0XBH9 M3X7J4 A0A195FJ62 F6VG59 A0A2M4CQE3 A0A1U7RPC2 A0A1S4JMR1 A0A3Q3AFG7 B4JAF1
A0A212EWA6 A0A2W1BEV1 A0A194PZ46 S4PE45 A0A1J1IPS7 D2A2L2 A0A336LU61 A0A0Q9WBD5 A0A0P8XV28 A0A034WAS0 A0A0K8VDI5 A0A3B0KEY2 A0A0Q9X5S1 A0A1L5L9D6 A0A0R3NXV6 A0A0A1WQ03 A0A1W4XHB0 W8BZB0 A0A0C9RLV6 M9ND31 A0A1B6F5M2 A0A0M4E313 A0A1W4UIY2 V5FZ40 A0A336LLF8 A0A0R1DSK6 A0A1I8MDQ5 A0A034WDD6 A0A0K8UEW5 A0A1B6H960 W8BS96 U5EY06 A0A0Q5WIN9 E0VTH1 A0A0J9TG94 A0A0A1XQ14 A4IHW6 B4LU11 A0A2J7PZD3 A0A0P8ZES1 B4KF38 A0A1L8E1Z9 A0A2P8Y487 A0A0P5S8V7 B3MMZ7 A0A0L0CAZ7 A0A0R3NRD4 A0A1B6DAT5 B4GJK4 A0A3B0K811 A0A1L8HTM5 Q6PA43 E9G0Q9 A0A1B6L607 A0A1I8PZ69 Q29NT9 D3TP29 A0A0P5XS92 A0A1B6KT34 A0A1I8MDR2 A0A1Y1L0K0 B0WKW4 A0A250YE98 A0A411G6A4 W5J8R2 A0A158P2B3 A0A3Q3J3M3 A0A2U3V426 A0A340WTF3 A0A2Y9MIC1 A0A384A0B1 A0A2Y9TFS2 A0A341AA49 Q16HW7-3 F1S3T0 A0A1S2ZYD2 A0A2M4BWA9 A0A2K5RPG0 A0A195D4E5 G3VGH7 A0A1B0D5D0 F4W6K8 A0A2M4ALC9 E2RH59 A0A3Q7UXB9 H0XBH9 M3X7J4 A0A195FJ62 F6VG59 A0A2M4CQE3 A0A1U7RPC2 A0A1S4JMR1 A0A3Q3AFG7 B4JAF1
EC Number
3.5.99.6
Pubmed
19121390
22118469
28756777
26354079
23622113
18362917
+ More
19820115 17994087 25348373 18057021 15632085 25830018 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25315136 20566863 22936249 29403074 26108605 27762356 21292972 20353571 28004739 28087693 20920257 23761445 21347285 17510324 21709235 21719571 16341006 17975172 19892987
19820115 17994087 25348373 18057021 15632085 25830018 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25315136 20566863 22936249 29403074 26108605 27762356 21292972 20353571 28004739 28087693 20920257 23761445 21347285 17510324 21709235 21719571 16341006 17975172 19892987
EMBL
BABH01023047
KY563981
KY609555
ASO76387.1
ASO76419.1
NWSH01000904
+ More
PCG73598.1 ODYU01004479 SOQ44391.1 RSAL01000178 RVE44994.1 PCG73596.1 AGBW02012057 OWR45751.1 KZ150093 PZC73659.1 KQ459585 KPI98313.1 GAIX01003331 JAA89229.1 CVRI01000057 CRL02243.1 KQ971338 EFA02005.2 UFQT01000004 SSX17188.1 CH940649 KRF81997.1 CH902620 KPU73189.1 GAKP01007168 JAC51784.1 GDHF01023457 GDHF01019871 GDHF01016833 GDHF01015310 JAI28857.1 JAI32443.1 JAI35481.1 JAI37004.1 OUUW01000006 SPP82168.1 CH933807 KRG03551.1 KU323592 APO36902.1 CH379058 KRT03651.1 GBXI01013697 JAD00595.1 GAMC01004537 JAC02019.1 GBYB01014337 JAG84104.1 AE014134 AFH03572.1 GECZ01024269 JAS45500.1 CP012523 ALC40116.1 GALX01005521 JAB62945.1 SSX17187.1 CM000157 KRJ98096.1 GAKP01007169 JAC51783.1 GDHF01027454 GDHF01023575 GDHF01012610 GDHF01003986 JAI24860.1 JAI28739.1 JAI39704.1 JAI48328.1 GECU01036516 JAS71190.1 GAMC01004538 JAC02018.1 GANO01002257 JAB57614.1 CH954177 KQS70185.1 DS235765 EEB16677.1 CM002910 KMY88320.1 GBXI01001337 JAD12955.1 BC135724 EDW65064.2 NEVH01020335 PNF21682.1 KPU73187.1 EDW13021.2 GFDF01001347 JAV12737.1 PYGN01000947 PSN39082.1 GDIQ01209118 GDIP01142908 JAK42607.1 JAL60806.1 EDV31022.1 KPU73186.1 JRES01000668 KNC29407.1 KRT03650.1 GEDC01029595 GEDC01025620 GEDC01019562 GEDC01014494 GEDC01013490 GEDC01001607 GEDC01000886 JAS07703.1 JAS11678.1 JAS17736.1 JAS22804.1 JAS23808.1 JAS35691.1 JAS36412.1 CH479184 EDW36820.1 SPP82169.1 CM004466 OCT99466.1 BC060461 GL732528 EFX86948.1 GEBQ01020892 GEBQ01001779 JAT19085.1 JAT38198.1 EZ423181 ADD19457.1 GDIP01068265 LRGB01001581 JAM35450.1 KZS11259.1 GEBQ01025416 JAT14561.1 GEZM01068174 JAV67114.1 DS231977 EDS30096.1 GFFW01002841 JAV41947.1 MH365611 QBB01420.1 ADMH02002094 ETN59265.1 ADTU01007051 CH478133 AEMK02000064 GGFJ01008216 MBW57357.1 KQ976885 KYN07304.1 AEFK01217617 AEFK01217618 AEFK01217619 AJVK01025142 GL887707 EGI70343.1 GGFK01008268 MBW41589.1 AAEX03009029 AAQR03112932 AAQR03112933 AANG04001605 KQ981523 KYN40302.1 GGFL01003378 MBW67556.1 CH916368 EDW03822.1
PCG73598.1 ODYU01004479 SOQ44391.1 RSAL01000178 RVE44994.1 PCG73596.1 AGBW02012057 OWR45751.1 KZ150093 PZC73659.1 KQ459585 KPI98313.1 GAIX01003331 JAA89229.1 CVRI01000057 CRL02243.1 KQ971338 EFA02005.2 UFQT01000004 SSX17188.1 CH940649 KRF81997.1 CH902620 KPU73189.1 GAKP01007168 JAC51784.1 GDHF01023457 GDHF01019871 GDHF01016833 GDHF01015310 JAI28857.1 JAI32443.1 JAI35481.1 JAI37004.1 OUUW01000006 SPP82168.1 CH933807 KRG03551.1 KU323592 APO36902.1 CH379058 KRT03651.1 GBXI01013697 JAD00595.1 GAMC01004537 JAC02019.1 GBYB01014337 JAG84104.1 AE014134 AFH03572.1 GECZ01024269 JAS45500.1 CP012523 ALC40116.1 GALX01005521 JAB62945.1 SSX17187.1 CM000157 KRJ98096.1 GAKP01007169 JAC51783.1 GDHF01027454 GDHF01023575 GDHF01012610 GDHF01003986 JAI24860.1 JAI28739.1 JAI39704.1 JAI48328.1 GECU01036516 JAS71190.1 GAMC01004538 JAC02018.1 GANO01002257 JAB57614.1 CH954177 KQS70185.1 DS235765 EEB16677.1 CM002910 KMY88320.1 GBXI01001337 JAD12955.1 BC135724 EDW65064.2 NEVH01020335 PNF21682.1 KPU73187.1 EDW13021.2 GFDF01001347 JAV12737.1 PYGN01000947 PSN39082.1 GDIQ01209118 GDIP01142908 JAK42607.1 JAL60806.1 EDV31022.1 KPU73186.1 JRES01000668 KNC29407.1 KRT03650.1 GEDC01029595 GEDC01025620 GEDC01019562 GEDC01014494 GEDC01013490 GEDC01001607 GEDC01000886 JAS07703.1 JAS11678.1 JAS17736.1 JAS22804.1 JAS23808.1 JAS35691.1 JAS36412.1 CH479184 EDW36820.1 SPP82169.1 CM004466 OCT99466.1 BC060461 GL732528 EFX86948.1 GEBQ01020892 GEBQ01001779 JAT19085.1 JAT38198.1 EZ423181 ADD19457.1 GDIP01068265 LRGB01001581 JAM35450.1 KZS11259.1 GEBQ01025416 JAT14561.1 GEZM01068174 JAV67114.1 DS231977 EDS30096.1 GFFW01002841 JAV41947.1 MH365611 QBB01420.1 ADMH02002094 ETN59265.1 ADTU01007051 CH478133 AEMK02000064 GGFJ01008216 MBW57357.1 KQ976885 KYN07304.1 AEFK01217617 AEFK01217618 AEFK01217619 AJVK01025142 GL887707 EGI70343.1 GGFK01008268 MBW41589.1 AAEX03009029 AAQR03112932 AAQR03112933 AANG04001605 KQ981523 KYN40302.1 GGFL01003378 MBW67556.1 CH916368 EDW03822.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000183832
+ More
UP000007266 UP000008792 UP000007801 UP000268350 UP000009192 UP000001819 UP000192223 UP000000803 UP000092553 UP000192221 UP000002282 UP000095301 UP000008711 UP000009046 UP000008143 UP000235965 UP000245037 UP000037069 UP000008744 UP000186698 UP000000305 UP000095300 UP000076858 UP000002320 UP000000673 UP000005205 UP000261600 UP000245320 UP000265300 UP000248483 UP000261681 UP000248484 UP000252040 UP000008820 UP000008227 UP000079721 UP000233040 UP000078542 UP000007648 UP000092462 UP000007755 UP000002254 UP000286640 UP000005225 UP000011712 UP000078541 UP000002281 UP000189705 UP000264800 UP000001070
UP000007266 UP000008792 UP000007801 UP000268350 UP000009192 UP000001819 UP000192223 UP000000803 UP000092553 UP000192221 UP000002282 UP000095301 UP000008711 UP000009046 UP000008143 UP000235965 UP000245037 UP000037069 UP000008744 UP000186698 UP000000305 UP000095300 UP000076858 UP000002320 UP000000673 UP000005205 UP000261600 UP000245320 UP000265300 UP000248483 UP000261681 UP000248484 UP000252040 UP000008820 UP000008227 UP000079721 UP000233040 UP000078542 UP000007648 UP000092462 UP000007755 UP000002254 UP000286640 UP000005225 UP000011712 UP000078541 UP000002281 UP000189705 UP000264800 UP000001070
PRIDE
Pfam
PF01182 Glucosamine_iso
Interpro
SUPFAM
SSF100950
SSF100950
ProteinModelPortal
H9JIW6
A0A222AJ35
A0A2A4JPG6
A0A2H1VU62
A0A437B3V5
A0A2A4JPB1
+ More
A0A212EWA6 A0A2W1BEV1 A0A194PZ46 S4PE45 A0A1J1IPS7 D2A2L2 A0A336LU61 A0A0Q9WBD5 A0A0P8XV28 A0A034WAS0 A0A0K8VDI5 A0A3B0KEY2 A0A0Q9X5S1 A0A1L5L9D6 A0A0R3NXV6 A0A0A1WQ03 A0A1W4XHB0 W8BZB0 A0A0C9RLV6 M9ND31 A0A1B6F5M2 A0A0M4E313 A0A1W4UIY2 V5FZ40 A0A336LLF8 A0A0R1DSK6 A0A1I8MDQ5 A0A034WDD6 A0A0K8UEW5 A0A1B6H960 W8BS96 U5EY06 A0A0Q5WIN9 E0VTH1 A0A0J9TG94 A0A0A1XQ14 A4IHW6 B4LU11 A0A2J7PZD3 A0A0P8ZES1 B4KF38 A0A1L8E1Z9 A0A2P8Y487 A0A0P5S8V7 B3MMZ7 A0A0L0CAZ7 A0A0R3NRD4 A0A1B6DAT5 B4GJK4 A0A3B0K811 A0A1L8HTM5 Q6PA43 E9G0Q9 A0A1B6L607 A0A1I8PZ69 Q29NT9 D3TP29 A0A0P5XS92 A0A1B6KT34 A0A1I8MDR2 A0A1Y1L0K0 B0WKW4 A0A250YE98 A0A411G6A4 W5J8R2 A0A158P2B3 A0A3Q3J3M3 A0A2U3V426 A0A340WTF3 A0A2Y9MIC1 A0A384A0B1 A0A2Y9TFS2 A0A341AA49 Q16HW7-3 F1S3T0 A0A1S2ZYD2 A0A2M4BWA9 A0A2K5RPG0 A0A195D4E5 G3VGH7 A0A1B0D5D0 F4W6K8 A0A2M4ALC9 E2RH59 A0A3Q7UXB9 H0XBH9 M3X7J4 A0A195FJ62 F6VG59 A0A2M4CQE3 A0A1U7RPC2 A0A1S4JMR1 A0A3Q3AFG7 B4JAF1
A0A212EWA6 A0A2W1BEV1 A0A194PZ46 S4PE45 A0A1J1IPS7 D2A2L2 A0A336LU61 A0A0Q9WBD5 A0A0P8XV28 A0A034WAS0 A0A0K8VDI5 A0A3B0KEY2 A0A0Q9X5S1 A0A1L5L9D6 A0A0R3NXV6 A0A0A1WQ03 A0A1W4XHB0 W8BZB0 A0A0C9RLV6 M9ND31 A0A1B6F5M2 A0A0M4E313 A0A1W4UIY2 V5FZ40 A0A336LLF8 A0A0R1DSK6 A0A1I8MDQ5 A0A034WDD6 A0A0K8UEW5 A0A1B6H960 W8BS96 U5EY06 A0A0Q5WIN9 E0VTH1 A0A0J9TG94 A0A0A1XQ14 A4IHW6 B4LU11 A0A2J7PZD3 A0A0P8ZES1 B4KF38 A0A1L8E1Z9 A0A2P8Y487 A0A0P5S8V7 B3MMZ7 A0A0L0CAZ7 A0A0R3NRD4 A0A1B6DAT5 B4GJK4 A0A3B0K811 A0A1L8HTM5 Q6PA43 E9G0Q9 A0A1B6L607 A0A1I8PZ69 Q29NT9 D3TP29 A0A0P5XS92 A0A1B6KT34 A0A1I8MDR2 A0A1Y1L0K0 B0WKW4 A0A250YE98 A0A411G6A4 W5J8R2 A0A158P2B3 A0A3Q3J3M3 A0A2U3V426 A0A340WTF3 A0A2Y9MIC1 A0A384A0B1 A0A2Y9TFS2 A0A341AA49 Q16HW7-3 F1S3T0 A0A1S2ZYD2 A0A2M4BWA9 A0A2K5RPG0 A0A195D4E5 G3VGH7 A0A1B0D5D0 F4W6K8 A0A2M4ALC9 E2RH59 A0A3Q7UXB9 H0XBH9 M3X7J4 A0A195FJ62 F6VG59 A0A2M4CQE3 A0A1U7RPC2 A0A1S4JMR1 A0A3Q3AFG7 B4JAF1
PDB
1NE7
E-value=1.11342e-112,
Score=1038
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
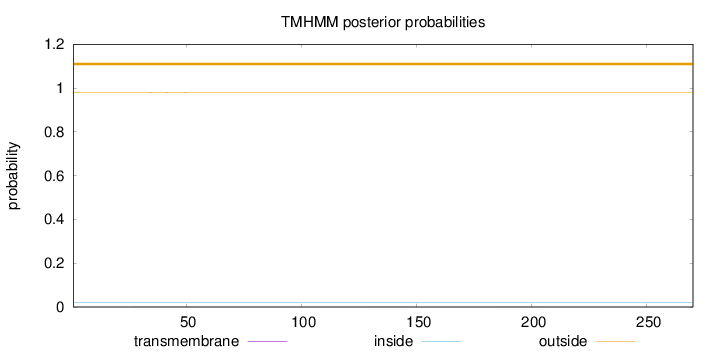
Length:
270
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02546
Exp number, first 60 AAs:
0.01629
Total prob of N-in:
0.01966
outside
1 - 270
Population Genetic Test Statistics
Pi
294.583983
Theta
184.80966
Tajima's D
1.598095
CLR
0.156018
CSRT
0.810159492025399
Interpretation
Uncertain
