Pre Gene Modal
BGIBMGA009495
Annotation
replication_factor_C_subunit_2_[Bombyx_mori]
Full name
Replication factor C subunit 2
Alternative Name
Activator 1 40 kDa subunit
Activator 1 subunit 2
Replication factor C 40 kDa subunit
Replication factor C subunit 4
Activator 1 subunit 2
Replication factor C 40 kDa subunit
Replication factor C subunit 4
Location in the cell
Cytoplasmic Reliability : 2.002
Sequence
CDS
ATGGATGATGATGATATCGTGATAATGGAAGTAGACGAACCGGAGAAGAAACAAGTAAAACCTAGTTCTAGTAAATCTACAAATTTACCATGGATAGAGAAATACAGGCCACAAACTTTCGACGATATCGTTGGCAATGAAGACACTGTGTCCCGCTTAGCCGTCTTTGCTAAAACCGGCAATGCTCCTAACATTATTATAGCTGGACCTCCTGGTGTTGGAAAAACGACGACTATACTATGCTTAGCGCGAGCTCTCCTCGGAGTATCTTTCAAAGACGCCGTATTAGAACTAAACGCCTCAAATGATCGTGGTATAGATGTCGTCCGGAATAAGATAAAAATGTTTGCGCAACAAAAGGTTACTTTGCCACCTGGTCGACATAAAATAGTGATACTAGATGAAGCGGACAGCATGACTGACGGAGCTCAACAAGCATTAAGACGCACCATGGAGCTGTATTCCAGTACAACTCGTTTTGCATTAGCAGCCAATAATAGTGAGAAGATAATAGAACCCATACAATCTAGGTGTGCTGTACTCCGGTACTCCAGGCTTAGTGATGCACAGATACTAGCAAAGGTTATAGAGATCTGCAATAAAGAAAACCTATCATACACTGAAGAAGGTGTGAGTGCAGTAGTCTTCACAGCACAGGGGGACTTGCGATCAGCCCTGAACAACCTGCAGTCAACAGCCCAGGGTTTTGGTCACATCAGCCCGGACAATGTGTTCAAAGTGTGTGATGAACCACATCCGATGGTGGTCAGGCAGATGCTTGAAGCTTGCACTAAACAGGATATACATGAGGCTTACAAGGTTATAGCAAAACTCTGCAAGATTGGCTACGCAGCAGAGGACATAGTCAGTAACATATTCAGAGTTTGCAAAACTTTAGATATATCTGAAGAGTTAAAACTGGCTTTCATCAGAGAGATTGGTTTGACACACATGAGGGTAGCAGATGGTCTTTCGTCTCCCCTGCAGCTGGCCGGTCTCCTCGCCAGGATGTGCAGGGTGGCTGCCGGCGGTGACCCGGAGTGGTGA
Protein
MDDDDIVIMEVDEPEKKQVKPSSSKSTNLPWIEKYRPQTFDDIVGNEDTVSRLAVFAKTGNAPNIIIAGPPGVGKTTTILCLARALLGVSFKDAVLELNASNDRGIDVVRNKIKMFAQQKVTLPPGRHKIVILDEADSMTDGAQQALRRTMELYSSTTRFALAANNSEKIIEPIQSRCAVLRYSRLSDAQILAKVIEICNKENLSYTEEGVSAVVFTAQGDLRSALNNLQSTAQGFGHISPDNVFKVCDEPHPMVVRQMLEACTKQDIHEAYKVIAKLCKIGYAAEDIVSNIFRVCKTLDISEELKLAFIREIGLTHMRVADGLSSPLQLAGLLARMCRVAAGGDPEW
Summary
Description
The elongation of primed DNA templates by DNA polymerase delta and epsilon requires the action of the accessory proteins proliferating cell nuclear antigen (PCNA) and activator 1. Subunit 2 binds ATP.
Subunit
Heteropentamer of subunits of 140/145, 40, 38, 37, and 36.5 kDa that forms a complex with PCNA in the presence of ATP.
Similarity
Belongs to the activator 1 small subunits family.
Keywords
ATP-binding
Cell cycle
Complete proteome
DNA replication
DNA-binding
Nucleotide-binding
Nucleus
Reference proteome
Feature
chain Replication factor C subunit 2
Uniprot
H9JIZ4
Q60GE7
A0A3S2LVP1
A0A0N0PBU5
A0A212EW79
A0A194PYA2
+ More
A0A2A4JNE6 A0A2W1BFU3 S4NT32 A0A1Q3EXN7 B0VZJ1 A0A182GXT3 A0A182JB41 A0A084WNA0 U5EV45 W5JRP3 A0A182FF42 A0A182MRS4 A0A182VDF2 A0A182KGH8 A0A182I3A5 A0A182KRC2 A0A182U182 A0A1I8MP82 A0A182S7T7 Q7QJE6 A0A182X3R4 A0A182QT42 A0A182XYW1 A0A0P6JS08 A0A0L7LAH0 A0A0L0C9Z6 A0A0T6B1B8 A0A1I8PJQ7 A0A336LWV6 A0A034W201 A0A0V0G645 A0A146LRR5 A0A146MDJ8 W8C2R8 N6UL65 R4FL91 A0A1A9V4P4 A0A0K8T2R4 A0A0K8UA74 A0A182PQF3 A0A154P8A8 A0A023F8X5 A0A1Y1LD99 A0A0A1WY16 A0A1L8DWT2 D3TS38 B4N3J7 B4L9D9 A0A1A9ZLX9 A0A1A9WAT6 A0A1B0C945 B4LB85 A0A195CFP7 B4J1E1 K7IMQ4 A0A182RSN2 A0A0L7RDR1 B4HU02 B4QQ45 A0A1W4W4I9 A0A0M4EZC3 A0A0N0U436 E2AG75 A0A2T7PCM7 P53034 B4PHP8 A0A310SQG9 A0A2J7QRU6 A0A232F9J5 A0A182WJ86 A0A0L8HHW0 A0A1B6KNQ8 A0A3B0JUP4 A0A1W4X0W3 E2BC66 B3NG38 A0A1B6EQ14 D7ELV8 A0A2R7W037 A0A0J7L2V6 A0A1S3DK53 A0A1B6C2G3 A0A195F9P0 F4X840 A0A402EMH7 A0A182NHV7 A0A067RQC0 A0A0N8AG98 B3M4V2 A0A164MZD4 A0A158NCB2 A0A452DYI2 E9IME3 A0A195BW97
A0A2A4JNE6 A0A2W1BFU3 S4NT32 A0A1Q3EXN7 B0VZJ1 A0A182GXT3 A0A182JB41 A0A084WNA0 U5EV45 W5JRP3 A0A182FF42 A0A182MRS4 A0A182VDF2 A0A182KGH8 A0A182I3A5 A0A182KRC2 A0A182U182 A0A1I8MP82 A0A182S7T7 Q7QJE6 A0A182X3R4 A0A182QT42 A0A182XYW1 A0A0P6JS08 A0A0L7LAH0 A0A0L0C9Z6 A0A0T6B1B8 A0A1I8PJQ7 A0A336LWV6 A0A034W201 A0A0V0G645 A0A146LRR5 A0A146MDJ8 W8C2R8 N6UL65 R4FL91 A0A1A9V4P4 A0A0K8T2R4 A0A0K8UA74 A0A182PQF3 A0A154P8A8 A0A023F8X5 A0A1Y1LD99 A0A0A1WY16 A0A1L8DWT2 D3TS38 B4N3J7 B4L9D9 A0A1A9ZLX9 A0A1A9WAT6 A0A1B0C945 B4LB85 A0A195CFP7 B4J1E1 K7IMQ4 A0A182RSN2 A0A0L7RDR1 B4HU02 B4QQ45 A0A1W4W4I9 A0A0M4EZC3 A0A0N0U436 E2AG75 A0A2T7PCM7 P53034 B4PHP8 A0A310SQG9 A0A2J7QRU6 A0A232F9J5 A0A182WJ86 A0A0L8HHW0 A0A1B6KNQ8 A0A3B0JUP4 A0A1W4X0W3 E2BC66 B3NG38 A0A1B6EQ14 D7ELV8 A0A2R7W037 A0A0J7L2V6 A0A1S3DK53 A0A1B6C2G3 A0A195F9P0 F4X840 A0A402EMH7 A0A182NHV7 A0A067RQC0 A0A0N8AG98 B3M4V2 A0A164MZD4 A0A158NCB2 A0A452DYI2 E9IME3 A0A195BW97
Pubmed
19121390
26354079
22118469
28756777
23622113
26483478
+ More
24438588 20920257 23761445 20966253 25315136 12364791 14747013 17210077 25244985 26999592 26227816 26108605 25348373 26823975 24495485 23537049 25474469 28004739 25830018 20353571 17994087 20075255 22936249 20798317 7789770 10731132 12537572 12537569 11438670 17550304 28648823 18362917 19820115 21719571 24845553 21347285 21282665
24438588 20920257 23761445 20966253 25315136 12364791 14747013 17210077 25244985 26999592 26227816 26108605 25348373 26823975 24495485 23537049 25474469 28004739 25830018 20353571 17994087 20075255 22936249 20798317 7789770 10731132 12537572 12537569 11438670 17550304 28648823 18362917 19820115 21719571 24845553 21347285 21282665
EMBL
BABH01023047
AB177621
BAD61055.1
RSAL01000178
RVE44995.1
KQ460889
+ More
KPJ11072.1 AGBW02012057 OWR45752.1 KQ459585 KPI98312.1 NWSH01000904 PCG73595.1 KZ150093 PZC73658.1 GAIX01013812 JAA78748.1 GFDL01015077 JAV19968.1 DS231814 EDS31728.1 JXUM01020780 KQ560581 KXJ81815.1 ATLV01024580 KE525352 KFB51694.1 GANO01002032 JAB57839.1 ADMH02000549 ETN65953.1 AXCM01005343 APCN01000204 AAAB01008807 EAA04621.4 AXCN02000379 GDUN01000788 JAN95131.1 JTDY01001946 KOB72472.1 JRES01000713 KNC29035.1 LJIG01016346 KRT80878.1 UFQT01000259 SSX22532.1 GAKP01010793 GAKP01010792 JAC48160.1 GECL01002928 JAP03196.1 GDHC01008824 JAQ09805.1 GDHC01000825 JAQ17804.1 GAMC01000323 JAC06233.1 APGK01019085 KB740092 KB632429 ENN81426.1 ERL95468.1 ACPB03001111 GAHY01002189 JAA75321.1 GBRD01006352 GBRD01006351 JAG59470.1 GDHF01028833 GDHF01026860 JAI23481.1 JAI25454.1 KQ434842 KZC08102.1 GBBI01000822 JAC17890.1 GEZM01063721 JAV68997.1 GBXI01010328 JAD03964.1 GFDF01003191 JAV10893.1 CCAG010000815 EZ424240 ADD20516.1 CH964095 EDW79202.1 CH933816 EDW17314.1 AJWK01001897 CH940647 EDW68649.1 KQ977873 KYM99126.1 CH916366 EDV95832.1 KQ414612 KOC69127.1 CH480817 EDW50423.1 CM000363 CM002912 EDX09165.1 KMY97508.1 CP012525 ALC44037.1 KQ435840 KOX71452.1 GL439266 EFN67566.1 PZQS01000004 PVD31174.1 U15967 AE014296 AY094829 CM000159 EDW93357.1 KQ761679 OAD57175.1 NEVH01011885 PNF31300.1 NNAY01000685 OXU26987.1 KQ418097 KOF88843.1 GEBQ01026907 JAT13070.1 OUUW01000002 SPP77429.1 GL447256 EFN86693.1 CH954178 EDV50867.1 GECZ01029779 JAS39990.1 KQ971431 EFA12420.2 KK854187 PTY12808.1 LBMM01001179 KMQ96769.1 GEDC01029586 JAS07712.1 KQ981727 KYN37096.1 GL888912 EGI57389.1 BDOT01000024 GCF45414.1 KK852529 KDR21944.1 GDIP01150353 JAJ73049.1 CH902618 EDV40526.1 LRGB01002914 KZS05513.1 ADTU01011781 LWLT01000031 GL764132 EFZ18260.1 KQ976396 KYM92857.1
KPJ11072.1 AGBW02012057 OWR45752.1 KQ459585 KPI98312.1 NWSH01000904 PCG73595.1 KZ150093 PZC73658.1 GAIX01013812 JAA78748.1 GFDL01015077 JAV19968.1 DS231814 EDS31728.1 JXUM01020780 KQ560581 KXJ81815.1 ATLV01024580 KE525352 KFB51694.1 GANO01002032 JAB57839.1 ADMH02000549 ETN65953.1 AXCM01005343 APCN01000204 AAAB01008807 EAA04621.4 AXCN02000379 GDUN01000788 JAN95131.1 JTDY01001946 KOB72472.1 JRES01000713 KNC29035.1 LJIG01016346 KRT80878.1 UFQT01000259 SSX22532.1 GAKP01010793 GAKP01010792 JAC48160.1 GECL01002928 JAP03196.1 GDHC01008824 JAQ09805.1 GDHC01000825 JAQ17804.1 GAMC01000323 JAC06233.1 APGK01019085 KB740092 KB632429 ENN81426.1 ERL95468.1 ACPB03001111 GAHY01002189 JAA75321.1 GBRD01006352 GBRD01006351 JAG59470.1 GDHF01028833 GDHF01026860 JAI23481.1 JAI25454.1 KQ434842 KZC08102.1 GBBI01000822 JAC17890.1 GEZM01063721 JAV68997.1 GBXI01010328 JAD03964.1 GFDF01003191 JAV10893.1 CCAG010000815 EZ424240 ADD20516.1 CH964095 EDW79202.1 CH933816 EDW17314.1 AJWK01001897 CH940647 EDW68649.1 KQ977873 KYM99126.1 CH916366 EDV95832.1 KQ414612 KOC69127.1 CH480817 EDW50423.1 CM000363 CM002912 EDX09165.1 KMY97508.1 CP012525 ALC44037.1 KQ435840 KOX71452.1 GL439266 EFN67566.1 PZQS01000004 PVD31174.1 U15967 AE014296 AY094829 CM000159 EDW93357.1 KQ761679 OAD57175.1 NEVH01011885 PNF31300.1 NNAY01000685 OXU26987.1 KQ418097 KOF88843.1 GEBQ01026907 JAT13070.1 OUUW01000002 SPP77429.1 GL447256 EFN86693.1 CH954178 EDV50867.1 GECZ01029779 JAS39990.1 KQ971431 EFA12420.2 KK854187 PTY12808.1 LBMM01001179 KMQ96769.1 GEDC01029586 JAS07712.1 KQ981727 KYN37096.1 GL888912 EGI57389.1 BDOT01000024 GCF45414.1 KK852529 KDR21944.1 GDIP01150353 JAJ73049.1 CH902618 EDV40526.1 LRGB01002914 KZS05513.1 ADTU01011781 LWLT01000031 GL764132 EFZ18260.1 KQ976396 KYM92857.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000053268
UP000218220
+ More
UP000002320 UP000069940 UP000249989 UP000075880 UP000030765 UP000000673 UP000069272 UP000075883 UP000075903 UP000075881 UP000075840 UP000075882 UP000075902 UP000095301 UP000075901 UP000007062 UP000076407 UP000075886 UP000076408 UP000037510 UP000037069 UP000095300 UP000019118 UP000030742 UP000015103 UP000078200 UP000075885 UP000076502 UP000092444 UP000007798 UP000009192 UP000092445 UP000091820 UP000092461 UP000008792 UP000078542 UP000001070 UP000002358 UP000075900 UP000053825 UP000001292 UP000000304 UP000192221 UP000092553 UP000053105 UP000000311 UP000245119 UP000000803 UP000002282 UP000235965 UP000215335 UP000075920 UP000053454 UP000268350 UP000192223 UP000008237 UP000008711 UP000007266 UP000036403 UP000079169 UP000078541 UP000007755 UP000288954 UP000075884 UP000027135 UP000007801 UP000076858 UP000005205 UP000291000 UP000078540
UP000002320 UP000069940 UP000249989 UP000075880 UP000030765 UP000000673 UP000069272 UP000075883 UP000075903 UP000075881 UP000075840 UP000075882 UP000075902 UP000095301 UP000075901 UP000007062 UP000076407 UP000075886 UP000076408 UP000037510 UP000037069 UP000095300 UP000019118 UP000030742 UP000015103 UP000078200 UP000075885 UP000076502 UP000092444 UP000007798 UP000009192 UP000092445 UP000091820 UP000092461 UP000008792 UP000078542 UP000001070 UP000002358 UP000075900 UP000053825 UP000001292 UP000000304 UP000192221 UP000092553 UP000053105 UP000000311 UP000245119 UP000000803 UP000002282 UP000235965 UP000215335 UP000075920 UP000053454 UP000268350 UP000192223 UP000008237 UP000008711 UP000007266 UP000036403 UP000079169 UP000078541 UP000007755 UP000288954 UP000075884 UP000027135 UP000007801 UP000076858 UP000005205 UP000291000 UP000078540
Interpro
ProteinModelPortal
H9JIZ4
Q60GE7
A0A3S2LVP1
A0A0N0PBU5
A0A212EW79
A0A194PYA2
+ More
A0A2A4JNE6 A0A2W1BFU3 S4NT32 A0A1Q3EXN7 B0VZJ1 A0A182GXT3 A0A182JB41 A0A084WNA0 U5EV45 W5JRP3 A0A182FF42 A0A182MRS4 A0A182VDF2 A0A182KGH8 A0A182I3A5 A0A182KRC2 A0A182U182 A0A1I8MP82 A0A182S7T7 Q7QJE6 A0A182X3R4 A0A182QT42 A0A182XYW1 A0A0P6JS08 A0A0L7LAH0 A0A0L0C9Z6 A0A0T6B1B8 A0A1I8PJQ7 A0A336LWV6 A0A034W201 A0A0V0G645 A0A146LRR5 A0A146MDJ8 W8C2R8 N6UL65 R4FL91 A0A1A9V4P4 A0A0K8T2R4 A0A0K8UA74 A0A182PQF3 A0A154P8A8 A0A023F8X5 A0A1Y1LD99 A0A0A1WY16 A0A1L8DWT2 D3TS38 B4N3J7 B4L9D9 A0A1A9ZLX9 A0A1A9WAT6 A0A1B0C945 B4LB85 A0A195CFP7 B4J1E1 K7IMQ4 A0A182RSN2 A0A0L7RDR1 B4HU02 B4QQ45 A0A1W4W4I9 A0A0M4EZC3 A0A0N0U436 E2AG75 A0A2T7PCM7 P53034 B4PHP8 A0A310SQG9 A0A2J7QRU6 A0A232F9J5 A0A182WJ86 A0A0L8HHW0 A0A1B6KNQ8 A0A3B0JUP4 A0A1W4X0W3 E2BC66 B3NG38 A0A1B6EQ14 D7ELV8 A0A2R7W037 A0A0J7L2V6 A0A1S3DK53 A0A1B6C2G3 A0A195F9P0 F4X840 A0A402EMH7 A0A182NHV7 A0A067RQC0 A0A0N8AG98 B3M4V2 A0A164MZD4 A0A158NCB2 A0A452DYI2 E9IME3 A0A195BW97
A0A2A4JNE6 A0A2W1BFU3 S4NT32 A0A1Q3EXN7 B0VZJ1 A0A182GXT3 A0A182JB41 A0A084WNA0 U5EV45 W5JRP3 A0A182FF42 A0A182MRS4 A0A182VDF2 A0A182KGH8 A0A182I3A5 A0A182KRC2 A0A182U182 A0A1I8MP82 A0A182S7T7 Q7QJE6 A0A182X3R4 A0A182QT42 A0A182XYW1 A0A0P6JS08 A0A0L7LAH0 A0A0L0C9Z6 A0A0T6B1B8 A0A1I8PJQ7 A0A336LWV6 A0A034W201 A0A0V0G645 A0A146LRR5 A0A146MDJ8 W8C2R8 N6UL65 R4FL91 A0A1A9V4P4 A0A0K8T2R4 A0A0K8UA74 A0A182PQF3 A0A154P8A8 A0A023F8X5 A0A1Y1LD99 A0A0A1WY16 A0A1L8DWT2 D3TS38 B4N3J7 B4L9D9 A0A1A9ZLX9 A0A1A9WAT6 A0A1B0C945 B4LB85 A0A195CFP7 B4J1E1 K7IMQ4 A0A182RSN2 A0A0L7RDR1 B4HU02 B4QQ45 A0A1W4W4I9 A0A0M4EZC3 A0A0N0U436 E2AG75 A0A2T7PCM7 P53034 B4PHP8 A0A310SQG9 A0A2J7QRU6 A0A232F9J5 A0A182WJ86 A0A0L8HHW0 A0A1B6KNQ8 A0A3B0JUP4 A0A1W4X0W3 E2BC66 B3NG38 A0A1B6EQ14 D7ELV8 A0A2R7W037 A0A0J7L2V6 A0A1S3DK53 A0A1B6C2G3 A0A195F9P0 F4X840 A0A402EMH7 A0A182NHV7 A0A067RQC0 A0A0N8AG98 B3M4V2 A0A164MZD4 A0A158NCB2 A0A452DYI2 E9IME3 A0A195BW97
PDB
1SXJ
E-value=1.1651e-105,
Score=978
Ontologies
KEGG
PATHWAY
GO
Topology
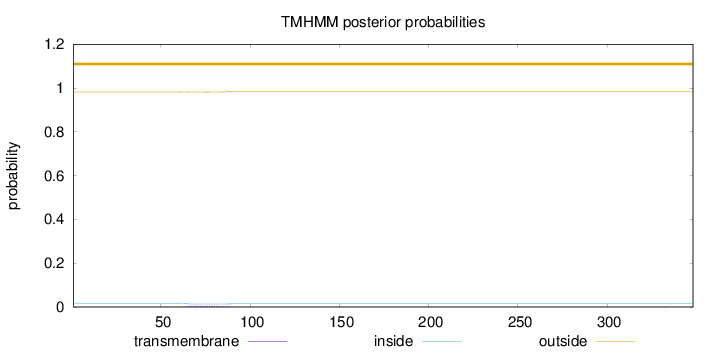
Subcellular location
Nucleus
Length:
348
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11457
Exp number, first 60 AAs:
0.00128
Total prob of N-in:
0.01769
outside
1 - 348
Population Genetic Test Statistics
Pi
184.985028
Theta
166.462806
Tajima's D
0.483134
CLR
0.164742
CSRT
0.508974551272436
Interpretation
Uncertain
