Pre Gene Modal
BGIBMGA009468
Annotation
PREDICTED:_charged_multivesicular_body_protein_2a_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.483
Sequence
CDS
ATGATGGAGTGGTTATTTGGTCACAAGATGACTCCAGATGAGATGCTGCGCAAGAATCAGCGAGCTCTCAACAAGGCCATGAGAGACCTGGACCGTGAACGAATGAAAATGGAACAGCAAGAGAAGAAGGTCATTGCTGACATTAAAAAGCTAGCTAAAGAAGGACAAATGGATGCTGTTAAAATCATGGCAAAAGACTTAGTACGTACCAGACGATATGTCCGCAAGTTCATGTTAATGAAAGCGAACATCCAAGCGGTGTCACTGAAGATACAAACTCTCAAGTCTCAGAGCACCATGGCGCAGGCGATGAAGGGAGTCACTCGTGCCATGGCTACTATGAATAGACAACTAAACATGCCACAGATACAAAAGATATTACAGGAGTTTGAAAAGCAGTCTGAAATTATGGATATGAAGGAGGAGATGATGAATGACTCCATCGATGAAGCGATGGAGGGAGACGATGATGAAGAAGAAAGTGACGCCGTCGTGTCACAGATCTTGGACGAGCTGGGCCTGCAGCTGAACGACACGCTGTCCGGGCTACCTCAGGCCACTGGATCCCTCTCTGTTGCCGCGAAGACCCCGGCGTCCCCGACGGCGGTGGCGGGCGGCGCCGGAGCCCCCGTGTCGGACGCGGACGCCGAGCTGCAGGCCCGCCTCGACAACCTGCGCCGGGAGTAG
Protein
MMEWLFGHKMTPDEMLRKNQRALNKAMRDLDRERMKMEQQEKKVIADIKKLAKEGQMDAVKIMAKDLVRTRRYVRKFMLMKANIQAVSLKIQTLKSQSTMAQAMKGVTRAMATMNRQLNMPQIQKILQEFEKQSEIMDMKEEMMNDSIDEAMEGDDDEEESDAVVSQILDELGLQLNDTLSGLPQATGSLSVAAKTPASPTAVAGGAGAPVSDADAELQARLDNLRRE
Summary
Uniprot
A0A2H1WXL2
A0A2W1BK58
H9JIW7
A0A1E1WH55
A0A0N0PBL2
A0A212EW93
+ More
A0A2A4JQA0 S4NWJ4 A0A437B3E5 A0A194PY96 A0A0J7KWQ8 A0A088A330 A0A026WLI1 A0A0N0BD81 A0A2A3EAS2 V9IC29 A0A154PG45 E9IW29 E2AM53 E2BJ37 U5EYT7 Q17LI1 V5H307 A0A182FRM6 A0A182GIB2 A0A195CMG9 A0A2M4BYX9 F4X0W9 A0A158N9F6 B0X7L4 A0A232EU63 K7J2S4 A0A2M4AH03 A0A1L8DW04 A0A182V834 A0A182U680 A0A182L8R5 A0A182X9I9 Q7PUC1 A0A182HKH2 A0A0L7QQQ2 A0A182PRQ0 A0A2R7VWL0 A0A182K523 A0A151JVQ5 A0A084VXS0 A0A182VVV2 A0A2M3Z5D1 A0A182RVR2 A0A182QN77 A0A182NQM1 W5J4Y8 A0A182J6S3 A0A195E1G7 A0A182LZ98 A0A182YBG0 A0A310S7J9 A0A1Q3FHI5 A0A023EK05 A0A2J7PCH8 A0A224XJG2 A0A069DXK9 A0A0P4VKY7 R4G5C3 A0A1W4XFM6 A0A2P8Z989 A0A0A9VQT9 D2A197 A0A1B6FYK0 A0A1B6IHK6 E0VBI0 J3JVP8 A0A1A9WF51 A0A1B6KZ58 B4NK38 D3TNP7 A0A1A9XEX2 A0A1B0BEV7 U4UBM3 B4GEF8 B4ICI7 A0A023F9M4 A0A0L0C5Y8 B4QW22 B3P6A4 Q9VBI3 A0A3B0K5U3 A0A1W4V901 Q296Z4 B4PSX0 W8CDQ0 A0A1A9VDZ4 A0A0M5IZZ6 B4JGJ5 A0A0K8USG6 A0A034W4Y8 A0A0A1X9F0 A0A1L8EEA0 N6U6V0 A0A1I8PBQ9
A0A2A4JQA0 S4NWJ4 A0A437B3E5 A0A194PY96 A0A0J7KWQ8 A0A088A330 A0A026WLI1 A0A0N0BD81 A0A2A3EAS2 V9IC29 A0A154PG45 E9IW29 E2AM53 E2BJ37 U5EYT7 Q17LI1 V5H307 A0A182FRM6 A0A182GIB2 A0A195CMG9 A0A2M4BYX9 F4X0W9 A0A158N9F6 B0X7L4 A0A232EU63 K7J2S4 A0A2M4AH03 A0A1L8DW04 A0A182V834 A0A182U680 A0A182L8R5 A0A182X9I9 Q7PUC1 A0A182HKH2 A0A0L7QQQ2 A0A182PRQ0 A0A2R7VWL0 A0A182K523 A0A151JVQ5 A0A084VXS0 A0A182VVV2 A0A2M3Z5D1 A0A182RVR2 A0A182QN77 A0A182NQM1 W5J4Y8 A0A182J6S3 A0A195E1G7 A0A182LZ98 A0A182YBG0 A0A310S7J9 A0A1Q3FHI5 A0A023EK05 A0A2J7PCH8 A0A224XJG2 A0A069DXK9 A0A0P4VKY7 R4G5C3 A0A1W4XFM6 A0A2P8Z989 A0A0A9VQT9 D2A197 A0A1B6FYK0 A0A1B6IHK6 E0VBI0 J3JVP8 A0A1A9WF51 A0A1B6KZ58 B4NK38 D3TNP7 A0A1A9XEX2 A0A1B0BEV7 U4UBM3 B4GEF8 B4ICI7 A0A023F9M4 A0A0L0C5Y8 B4QW22 B3P6A4 Q9VBI3 A0A3B0K5U3 A0A1W4V901 Q296Z4 B4PSX0 W8CDQ0 A0A1A9VDZ4 A0A0M5IZZ6 B4JGJ5 A0A0K8USG6 A0A034W4Y8 A0A0A1X9F0 A0A1L8EEA0 N6U6V0 A0A1I8PBQ9
Pubmed
28756777
19121390
26354079
22118469
23622113
24508170
+ More
30249741 21282665 20798317 17510324 26483478 21719571 21347285 28648823 20075255 20966253 12364791 14747013 17210077 24438588 20920257 23761445 25244985 24945155 26334808 27129103 29403074 25401762 26823975 18362917 19820115 20566863 22516182 17994087 20353571 23537049 25474469 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 24495485 25348373 25830018
30249741 21282665 20798317 17510324 26483478 21719571 21347285 28648823 20075255 20966253 12364791 14747013 17210077 24438588 20920257 23761445 25244985 24945155 26334808 27129103 29403074 25401762 26823975 18362917 19820115 20566863 22516182 17994087 20353571 23537049 25474469 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 24495485 25348373 25830018
EMBL
ODYU01011840
SOQ57813.1
KZ150093
PZC73657.1
BABH01023047
GDQN01004863
+ More
JAT86191.1 KQ460889 KPJ11071.1 AGBW02012057 OWR45753.1 NWSH01000904 PCG73592.1 GAIX01012517 JAA80043.1 RSAL01000178 RVE44996.1 KQ459585 KPI98311.1 LBMM01002501 KMQ94733.1 KK107154 QOIP01000001 EZA56808.1 RLU27166.1 KQ435878 KOX70068.1 KZ288322 PBC28161.1 JR039394 AEY58673.1 KQ434896 KZC10802.1 GL766449 EFZ15208.1 GL440706 EFN65486.1 GL448558 EFN84187.1 GANO01000131 JAB59740.1 CH477215 EAT47555.1 GALX01001238 JAB67228.1 JXUM01065565 KQ562354 KXJ76092.1 KQ977565 KYN01921.1 GGFJ01008980 MBW58121.1 GL888503 EGI59911.1 ADTU01009631 DS232454 EDS42026.1 NNAY01002172 OXU21874.1 GGFK01006733 MBW40054.1 GFDF01003491 JAV10593.1 AAAB01008987 EAA01777.4 APCN01000830 KQ414786 KOC60879.1 KK854135 PTY11893.1 KQ981713 KYN37381.1 ATLV01018205 KE525224 KFB42764.1 GGFM01002976 MBW23727.1 AXCN02001514 ADMH02002105 ETN59026.1 KQ979814 KYN18973.1 AXCM01000928 KQ765605 OAD53847.1 GFDL01008019 JAV27026.1 GAPW01004162 JAC09436.1 NEVH01027059 PNF14034.1 GFTR01003800 JAW12626.1 GBGD01002800 JAC86089.1 GDKW01000822 JAI55773.1 GAHY01000665 JAA76845.1 PYGN01000141 PSN53063.1 GBHO01044802 GBRD01011504 GDHC01015497 JAF98801.1 JAG54320.1 JAQ03132.1 KQ971338 EFA01563.1 GECZ01020734 GECZ01020070 GECZ01020034 GECZ01014511 GECZ01003461 JAS49035.1 JAS49699.1 JAS49735.1 JAS55258.1 JAS66308.1 GECU01021313 JAS86393.1 DS235030 EEB10736.1 BT127316 AEE62278.1 GEBQ01026314 GEBQ01023250 GEBQ01017796 GEBQ01015611 GEBQ01011774 GEBQ01011753 GEBQ01009681 GEBQ01001530 JAT13663.1 JAT16727.1 JAT22181.1 JAT24366.1 JAT28203.1 JAT28224.1 JAT30296.1 JAT38447.1 CH964272 EDW85080.1 EZ423049 ADD19325.1 JXJN01013032 KB632012 ERL88001.1 CH479182 EDW33993.1 CH480828 EDW45083.1 GBBI01000535 JAC18177.1 JRES01000953 KNC26829.1 CM000364 EDX14521.1 CH954182 EDV53574.1 AE014297 BT003747 AAF56559.1 AAO41408.1 OUUW01000023 SPP89574.1 CM000070 EAL28312.2 CM000160 EDW98657.1 GAMC01001561 JAC04995.1 CP012526 ALC46736.1 CH916369 EDV93692.1 GDHF01023044 JAI29270.1 GAKP01010124 JAC48828.1 GBXI01006957 JAD07335.1 GFDG01001771 JAV17028.1 APGK01040441 KB740981 ENN76371.1
JAT86191.1 KQ460889 KPJ11071.1 AGBW02012057 OWR45753.1 NWSH01000904 PCG73592.1 GAIX01012517 JAA80043.1 RSAL01000178 RVE44996.1 KQ459585 KPI98311.1 LBMM01002501 KMQ94733.1 KK107154 QOIP01000001 EZA56808.1 RLU27166.1 KQ435878 KOX70068.1 KZ288322 PBC28161.1 JR039394 AEY58673.1 KQ434896 KZC10802.1 GL766449 EFZ15208.1 GL440706 EFN65486.1 GL448558 EFN84187.1 GANO01000131 JAB59740.1 CH477215 EAT47555.1 GALX01001238 JAB67228.1 JXUM01065565 KQ562354 KXJ76092.1 KQ977565 KYN01921.1 GGFJ01008980 MBW58121.1 GL888503 EGI59911.1 ADTU01009631 DS232454 EDS42026.1 NNAY01002172 OXU21874.1 GGFK01006733 MBW40054.1 GFDF01003491 JAV10593.1 AAAB01008987 EAA01777.4 APCN01000830 KQ414786 KOC60879.1 KK854135 PTY11893.1 KQ981713 KYN37381.1 ATLV01018205 KE525224 KFB42764.1 GGFM01002976 MBW23727.1 AXCN02001514 ADMH02002105 ETN59026.1 KQ979814 KYN18973.1 AXCM01000928 KQ765605 OAD53847.1 GFDL01008019 JAV27026.1 GAPW01004162 JAC09436.1 NEVH01027059 PNF14034.1 GFTR01003800 JAW12626.1 GBGD01002800 JAC86089.1 GDKW01000822 JAI55773.1 GAHY01000665 JAA76845.1 PYGN01000141 PSN53063.1 GBHO01044802 GBRD01011504 GDHC01015497 JAF98801.1 JAG54320.1 JAQ03132.1 KQ971338 EFA01563.1 GECZ01020734 GECZ01020070 GECZ01020034 GECZ01014511 GECZ01003461 JAS49035.1 JAS49699.1 JAS49735.1 JAS55258.1 JAS66308.1 GECU01021313 JAS86393.1 DS235030 EEB10736.1 BT127316 AEE62278.1 GEBQ01026314 GEBQ01023250 GEBQ01017796 GEBQ01015611 GEBQ01011774 GEBQ01011753 GEBQ01009681 GEBQ01001530 JAT13663.1 JAT16727.1 JAT22181.1 JAT24366.1 JAT28203.1 JAT28224.1 JAT30296.1 JAT38447.1 CH964272 EDW85080.1 EZ423049 ADD19325.1 JXJN01013032 KB632012 ERL88001.1 CH479182 EDW33993.1 CH480828 EDW45083.1 GBBI01000535 JAC18177.1 JRES01000953 KNC26829.1 CM000364 EDX14521.1 CH954182 EDV53574.1 AE014297 BT003747 AAF56559.1 AAO41408.1 OUUW01000023 SPP89574.1 CM000070 EAL28312.2 CM000160 EDW98657.1 GAMC01001561 JAC04995.1 CP012526 ALC46736.1 CH916369 EDV93692.1 GDHF01023044 JAI29270.1 GAKP01010124 JAC48828.1 GBXI01006957 JAD07335.1 GFDG01001771 JAV17028.1 APGK01040441 KB740981 ENN76371.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000283053
UP000053268
+ More
UP000036403 UP000005203 UP000053097 UP000279307 UP000053105 UP000242457 UP000076502 UP000000311 UP000008237 UP000008820 UP000069272 UP000069940 UP000249989 UP000078542 UP000007755 UP000005205 UP000002320 UP000215335 UP000002358 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000053825 UP000075885 UP000075881 UP000078541 UP000030765 UP000075920 UP000075900 UP000075886 UP000075884 UP000000673 UP000075880 UP000078492 UP000075883 UP000076408 UP000235965 UP000192223 UP000245037 UP000007266 UP000009046 UP000091820 UP000007798 UP000092443 UP000092460 UP000030742 UP000008744 UP000001292 UP000037069 UP000000304 UP000008711 UP000000803 UP000268350 UP000192221 UP000001819 UP000002282 UP000078200 UP000092553 UP000001070 UP000019118 UP000095300
UP000036403 UP000005203 UP000053097 UP000279307 UP000053105 UP000242457 UP000076502 UP000000311 UP000008237 UP000008820 UP000069272 UP000069940 UP000249989 UP000078542 UP000007755 UP000005205 UP000002320 UP000215335 UP000002358 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000053825 UP000075885 UP000075881 UP000078541 UP000030765 UP000075920 UP000075900 UP000075886 UP000075884 UP000000673 UP000075880 UP000078492 UP000075883 UP000076408 UP000235965 UP000192223 UP000245037 UP000007266 UP000009046 UP000091820 UP000007798 UP000092443 UP000092460 UP000030742 UP000008744 UP000001292 UP000037069 UP000000304 UP000008711 UP000000803 UP000268350 UP000192221 UP000001819 UP000002282 UP000078200 UP000092553 UP000001070 UP000019118 UP000095300
PRIDE
Pfam
PF03357 Snf7
Interpro
IPR005024
Snf7_fam
ProteinModelPortal
A0A2H1WXL2
A0A2W1BK58
H9JIW7
A0A1E1WH55
A0A0N0PBL2
A0A212EW93
+ More
A0A2A4JQA0 S4NWJ4 A0A437B3E5 A0A194PY96 A0A0J7KWQ8 A0A088A330 A0A026WLI1 A0A0N0BD81 A0A2A3EAS2 V9IC29 A0A154PG45 E9IW29 E2AM53 E2BJ37 U5EYT7 Q17LI1 V5H307 A0A182FRM6 A0A182GIB2 A0A195CMG9 A0A2M4BYX9 F4X0W9 A0A158N9F6 B0X7L4 A0A232EU63 K7J2S4 A0A2M4AH03 A0A1L8DW04 A0A182V834 A0A182U680 A0A182L8R5 A0A182X9I9 Q7PUC1 A0A182HKH2 A0A0L7QQQ2 A0A182PRQ0 A0A2R7VWL0 A0A182K523 A0A151JVQ5 A0A084VXS0 A0A182VVV2 A0A2M3Z5D1 A0A182RVR2 A0A182QN77 A0A182NQM1 W5J4Y8 A0A182J6S3 A0A195E1G7 A0A182LZ98 A0A182YBG0 A0A310S7J9 A0A1Q3FHI5 A0A023EK05 A0A2J7PCH8 A0A224XJG2 A0A069DXK9 A0A0P4VKY7 R4G5C3 A0A1W4XFM6 A0A2P8Z989 A0A0A9VQT9 D2A197 A0A1B6FYK0 A0A1B6IHK6 E0VBI0 J3JVP8 A0A1A9WF51 A0A1B6KZ58 B4NK38 D3TNP7 A0A1A9XEX2 A0A1B0BEV7 U4UBM3 B4GEF8 B4ICI7 A0A023F9M4 A0A0L0C5Y8 B4QW22 B3P6A4 Q9VBI3 A0A3B0K5U3 A0A1W4V901 Q296Z4 B4PSX0 W8CDQ0 A0A1A9VDZ4 A0A0M5IZZ6 B4JGJ5 A0A0K8USG6 A0A034W4Y8 A0A0A1X9F0 A0A1L8EEA0 N6U6V0 A0A1I8PBQ9
A0A2A4JQA0 S4NWJ4 A0A437B3E5 A0A194PY96 A0A0J7KWQ8 A0A088A330 A0A026WLI1 A0A0N0BD81 A0A2A3EAS2 V9IC29 A0A154PG45 E9IW29 E2AM53 E2BJ37 U5EYT7 Q17LI1 V5H307 A0A182FRM6 A0A182GIB2 A0A195CMG9 A0A2M4BYX9 F4X0W9 A0A158N9F6 B0X7L4 A0A232EU63 K7J2S4 A0A2M4AH03 A0A1L8DW04 A0A182V834 A0A182U680 A0A182L8R5 A0A182X9I9 Q7PUC1 A0A182HKH2 A0A0L7QQQ2 A0A182PRQ0 A0A2R7VWL0 A0A182K523 A0A151JVQ5 A0A084VXS0 A0A182VVV2 A0A2M3Z5D1 A0A182RVR2 A0A182QN77 A0A182NQM1 W5J4Y8 A0A182J6S3 A0A195E1G7 A0A182LZ98 A0A182YBG0 A0A310S7J9 A0A1Q3FHI5 A0A023EK05 A0A2J7PCH8 A0A224XJG2 A0A069DXK9 A0A0P4VKY7 R4G5C3 A0A1W4XFM6 A0A2P8Z989 A0A0A9VQT9 D2A197 A0A1B6FYK0 A0A1B6IHK6 E0VBI0 J3JVP8 A0A1A9WF51 A0A1B6KZ58 B4NK38 D3TNP7 A0A1A9XEX2 A0A1B0BEV7 U4UBM3 B4GEF8 B4ICI7 A0A023F9M4 A0A0L0C5Y8 B4QW22 B3P6A4 Q9VBI3 A0A3B0K5U3 A0A1W4V901 Q296Z4 B4PSX0 W8CDQ0 A0A1A9VDZ4 A0A0M5IZZ6 B4JGJ5 A0A0K8USG6 A0A034W4Y8 A0A0A1X9F0 A0A1L8EEA0 N6U6V0 A0A1I8PBQ9
PDB
6E8G
E-value=3.74728e-10,
Score=152
Ontologies
GO
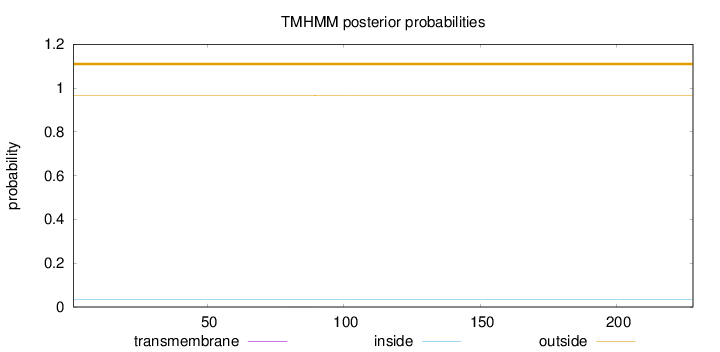
Topology
Length:
228
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00298
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03327
outside
1 - 228
Population Genetic Test Statistics
Pi
213.304822
Theta
185.171015
Tajima's D
0.491096
CLR
0.071005
CSRT
0.506924653767312
Interpretation
Uncertain
