Gene
KWMTBOMO08198
Pre Gene Modal
BGIBMGA009494
Annotation
PREDICTED:_lysoplasmalogenase-like_protein_TMEM86A_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.629
Sequence
CDS
ATGATTTCCCCTACTGCATTGGTAAAAAGGGTGGGGGTGGGCGGTCGTCTCGTCCCATTCTTCAAATGTGTCTGCGTGTATTTCCTGGCCATGGGCGCGGGTGGGAGCGATCCATCCTTGATCGGCGTGGCAACAAAATGCGCTCCGATACTTTGCCTGCTACTCGTCGTGGTTCTACACCATGGATCCTTGCCTTATGAGAAAGGTTGGTACGCGCGACGGATCGCGGCCGGTCTGGCTCTATCAGCCCTGGGCGATGCCTTCCTGGTTTGGCCGCAGCACTTCATCGCGGGCATGGCGGCCTTTGCGGCTGCCCACATATCCTACATATCAGCTTTCCGGTTCACCCCGCTCGCGAAGCCATTAGCACCTATCGTCTACTCGGGTTGCGGGTACTACATGACCTTGATAACTCCGCCTGATGGATTGAAGGTGTTGGTACCGGTGTACGGAATGTTATTGAGTACAATGGCATGGAGGGGCCTGGCCAGGGGTCGGGCGGCGGCTGTCGGTGCTGTACTGTTCCTGATATCTGATGCTATACTAGGGTACAGCCTATTTGGAGGACCGTAG
Protein
MISPTALVKRVGVGGRLVPFFKCVCVYFLAMGAGGSDPSLIGVATKCAPILCLLLVVVLHHGSLPYEKGWYARRIAAGLALSALGDAFLVWPQHFIAGMAAFAAAHISYISAFRFTPLAKPLAPIVYSGCGYYMTLITPPDGLKVLVPVYGMLLSTMAWRGLARGRAAAVGAVLFLISDAILGYSLFGGP
Summary
Uniprot
H9JIZ3
A0A212EWD2
A0A154PHS0
A0A310SC69
A0A0M8ZY85
A0A2A3E524
+ More
A0A087ZY52 A0A0L7R0P4 A0A3L8DID6 A0A195FR20 A0A158NA31 E9IXL7 F4WC27 K7JCT3 E2A5R4 E2B880 A0A1B6CW54 A0A0P5YDU3 E9FWP2 A0A0P5GT30 A0A0P5Y904 A0A0N8ASQ6 A0A0P6DPW4 A0A0P4ZE17 A0A0P5N1Y3 A0A0P5MIW0 A0A0P5AC23 A0A0P5AE42 A0A0P6AWR2 A0A0P6HG00 A0A2J7PSE3 A0A2Y9P3X3 A0A2Y9P6P5 A0A0P6AZL6 A0A0N8B0Y1 A0A2Y9P929 A0A2Y9P6P0 A0A0P4ZZL8 A0A341C807 A0A067QX94 A0A384A2I6 A0A341C803 A0A384A261 A0A0P5NBP6 A0A0P5YHT1 A0A0N8EC92 A0A0N1PIB9 S9XKE0 A0A1B6K172 A0A340Y8G3 A0A340Y494 T1JLT3 A0A0P5TQJ4 A0A1B6GGQ2 A0A2Y9R999 A0A444TRS5 A0A1B6LWI5 A0A1B6MS05 A0A2Y9EKA4 A0A2Y9PEU7 A0A2Y9PGH7 A0A2Y9EI79 F7AFA1 A0A139WIV7 A0A293M2S3
A0A087ZY52 A0A0L7R0P4 A0A3L8DID6 A0A195FR20 A0A158NA31 E9IXL7 F4WC27 K7JCT3 E2A5R4 E2B880 A0A1B6CW54 A0A0P5YDU3 E9FWP2 A0A0P5GT30 A0A0P5Y904 A0A0N8ASQ6 A0A0P6DPW4 A0A0P4ZE17 A0A0P5N1Y3 A0A0P5MIW0 A0A0P5AC23 A0A0P5AE42 A0A0P6AWR2 A0A0P6HG00 A0A2J7PSE3 A0A2Y9P3X3 A0A2Y9P6P5 A0A0P6AZL6 A0A0N8B0Y1 A0A2Y9P929 A0A2Y9P6P0 A0A0P4ZZL8 A0A341C807 A0A067QX94 A0A384A2I6 A0A341C803 A0A384A261 A0A0P5NBP6 A0A0P5YHT1 A0A0N8EC92 A0A0N1PIB9 S9XKE0 A0A1B6K172 A0A340Y8G3 A0A340Y494 T1JLT3 A0A0P5TQJ4 A0A1B6GGQ2 A0A2Y9R999 A0A444TRS5 A0A1B6LWI5 A0A1B6MS05 A0A2Y9EKA4 A0A2Y9PEU7 A0A2Y9PGH7 A0A2Y9EI79 F7AFA1 A0A139WIV7 A0A293M2S3
Pubmed
EMBL
BABH01023044
BABH01023045
AGBW02012057
OWR45754.1
KQ434912
KZC11373.1
+ More
KQ769812 OAD52843.1 KQ435826 KOX72019.1 KZ288369 PBC26788.1 KQ414670 KOC64408.1 QOIP01000007 RLU20204.1 KQ981305 KYN42918.1 ADTU01009955 GL766762 EFZ14627.1 GL888070 EGI68007.1 AAZX01014685 GL437023 EFN71261.1 GL446286 EFN88082.1 GEDC01024980 GEDC01023668 GEDC01019684 GEDC01006691 GEDC01004536 JAS12318.1 JAS13630.1 JAS17614.1 JAS30607.1 JAS32762.1 GDIP01059221 JAM44494.1 GL732526 EFX88398.1 GDIQ01236060 GDIQ01116352 JAK15665.1 GDIP01061168 JAM42547.1 GDIQ01246611 JAK05114.1 GDIQ01075415 JAN19322.1 GDIP01218299 JAJ05103.1 GDIQ01155989 JAK95736.1 GDIQ01155988 JAK95737.1 GDIP01205645 JAJ17757.1 GDIP01203447 JAJ19955.1 GDIP01029859 JAM73856.1 GDIQ01019978 JAN74759.1 NEVH01021931 PNF19257.1 GDIP01022091 JAM81624.1 GDIQ01223611 JAK28114.1 GDIP01209253 JAJ14149.1 KK852907 KDR13993.1 GDIQ01144393 JAL07333.1 GDIP01057513 JAM46202.1 GDIQ01040932 JAN53805.1 KQ460889 KPJ11070.1 KB016491 EPY88134.1 GECU01002497 JAT05210.1 JH432130 GDIP01123010 JAL80704.1 GECZ01008145 JAS61624.1 SAUD01000821 RXG72631.1 GEBQ01027304 GEBQ01011926 JAT12673.1 JAT28051.1 GEBQ01010493 GEBQ01001266 JAT29484.1 JAT38711.1 JSUE03022357 KQ971338 KYB27846.1 GFWV01010544 MAA35273.1
KQ769812 OAD52843.1 KQ435826 KOX72019.1 KZ288369 PBC26788.1 KQ414670 KOC64408.1 QOIP01000007 RLU20204.1 KQ981305 KYN42918.1 ADTU01009955 GL766762 EFZ14627.1 GL888070 EGI68007.1 AAZX01014685 GL437023 EFN71261.1 GL446286 EFN88082.1 GEDC01024980 GEDC01023668 GEDC01019684 GEDC01006691 GEDC01004536 JAS12318.1 JAS13630.1 JAS17614.1 JAS30607.1 JAS32762.1 GDIP01059221 JAM44494.1 GL732526 EFX88398.1 GDIQ01236060 GDIQ01116352 JAK15665.1 GDIP01061168 JAM42547.1 GDIQ01246611 JAK05114.1 GDIQ01075415 JAN19322.1 GDIP01218299 JAJ05103.1 GDIQ01155989 JAK95736.1 GDIQ01155988 JAK95737.1 GDIP01205645 JAJ17757.1 GDIP01203447 JAJ19955.1 GDIP01029859 JAM73856.1 GDIQ01019978 JAN74759.1 NEVH01021931 PNF19257.1 GDIP01022091 JAM81624.1 GDIQ01223611 JAK28114.1 GDIP01209253 JAJ14149.1 KK852907 KDR13993.1 GDIQ01144393 JAL07333.1 GDIP01057513 JAM46202.1 GDIQ01040932 JAN53805.1 KQ460889 KPJ11070.1 KB016491 EPY88134.1 GECU01002497 JAT05210.1 JH432130 GDIP01123010 JAL80704.1 GECZ01008145 JAS61624.1 SAUD01000821 RXG72631.1 GEBQ01027304 GEBQ01011926 JAT12673.1 JAT28051.1 GEBQ01010493 GEBQ01001266 JAT29484.1 JAT38711.1 JSUE03022357 KQ971338 KYB27846.1 GFWV01010544 MAA35273.1
Proteomes
UP000005204
UP000007151
UP000076502
UP000053105
UP000242457
UP000005203
+ More
UP000053825 UP000279307 UP000078541 UP000005205 UP000007755 UP000002358 UP000000311 UP000008237 UP000000305 UP000235965 UP000248483 UP000252040 UP000027135 UP000261681 UP000053240 UP000265300 UP000248480 UP000288706 UP000248484 UP000006718 UP000007266
UP000053825 UP000279307 UP000078541 UP000005205 UP000007755 UP000002358 UP000000311 UP000008237 UP000000305 UP000235965 UP000248483 UP000252040 UP000027135 UP000261681 UP000053240 UP000265300 UP000248480 UP000288706 UP000248484 UP000006718 UP000007266
PRIDE
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9JIZ3
A0A212EWD2
A0A154PHS0
A0A310SC69
A0A0M8ZY85
A0A2A3E524
+ More
A0A087ZY52 A0A0L7R0P4 A0A3L8DID6 A0A195FR20 A0A158NA31 E9IXL7 F4WC27 K7JCT3 E2A5R4 E2B880 A0A1B6CW54 A0A0P5YDU3 E9FWP2 A0A0P5GT30 A0A0P5Y904 A0A0N8ASQ6 A0A0P6DPW4 A0A0P4ZE17 A0A0P5N1Y3 A0A0P5MIW0 A0A0P5AC23 A0A0P5AE42 A0A0P6AWR2 A0A0P6HG00 A0A2J7PSE3 A0A2Y9P3X3 A0A2Y9P6P5 A0A0P6AZL6 A0A0N8B0Y1 A0A2Y9P929 A0A2Y9P6P0 A0A0P4ZZL8 A0A341C807 A0A067QX94 A0A384A2I6 A0A341C803 A0A384A261 A0A0P5NBP6 A0A0P5YHT1 A0A0N8EC92 A0A0N1PIB9 S9XKE0 A0A1B6K172 A0A340Y8G3 A0A340Y494 T1JLT3 A0A0P5TQJ4 A0A1B6GGQ2 A0A2Y9R999 A0A444TRS5 A0A1B6LWI5 A0A1B6MS05 A0A2Y9EKA4 A0A2Y9PEU7 A0A2Y9PGH7 A0A2Y9EI79 F7AFA1 A0A139WIV7 A0A293M2S3
A0A087ZY52 A0A0L7R0P4 A0A3L8DID6 A0A195FR20 A0A158NA31 E9IXL7 F4WC27 K7JCT3 E2A5R4 E2B880 A0A1B6CW54 A0A0P5YDU3 E9FWP2 A0A0P5GT30 A0A0P5Y904 A0A0N8ASQ6 A0A0P6DPW4 A0A0P4ZE17 A0A0P5N1Y3 A0A0P5MIW0 A0A0P5AC23 A0A0P5AE42 A0A0P6AWR2 A0A0P6HG00 A0A2J7PSE3 A0A2Y9P3X3 A0A2Y9P6P5 A0A0P6AZL6 A0A0N8B0Y1 A0A2Y9P929 A0A2Y9P6P0 A0A0P4ZZL8 A0A341C807 A0A067QX94 A0A384A2I6 A0A341C803 A0A384A261 A0A0P5NBP6 A0A0P5YHT1 A0A0N8EC92 A0A0N1PIB9 S9XKE0 A0A1B6K172 A0A340Y8G3 A0A340Y494 T1JLT3 A0A0P5TQJ4 A0A1B6GGQ2 A0A2Y9R999 A0A444TRS5 A0A1B6LWI5 A0A1B6MS05 A0A2Y9EKA4 A0A2Y9PEU7 A0A2Y9PGH7 A0A2Y9EI79 F7AFA1 A0A139WIV7 A0A293M2S3
Ontologies
GO
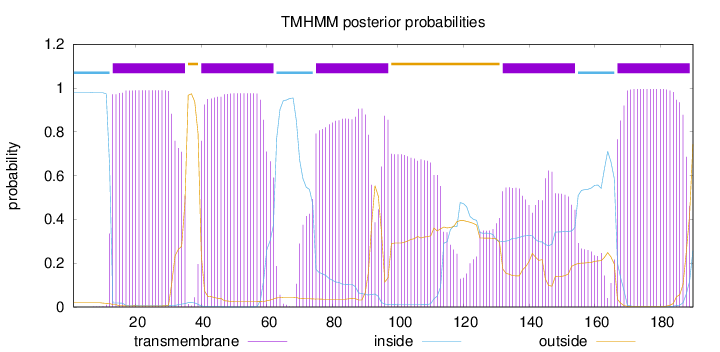
Topology
Length:
190
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
114.89115
Exp number, first 60 AAs:
41.62609
Total prob of N-in:
0.98026
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 39
TMhelix
40 - 62
inside
63 - 74
TMhelix
75 - 97
outside
98 - 131
TMhelix
132 - 154
inside
155 - 166
TMhelix
167 - 189
outside
190 - 190
Population Genetic Test Statistics
Pi
23.204584
Theta
187.437229
Tajima's D
0.696038
CLR
0.411238
CSRT
0.569221538923054
Interpretation
Uncertain
