Gene
KWMTBOMO08196
Pre Gene Modal
BGIBMGA009471
Annotation
PREDICTED:_rho_GTPase-activating_protein_26_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.14
Sequence
CDS
ATGGGCGTGGGTCTGCAGCCTCTAGAGTTCACCGAGTGCCTCGCGGACAGCCCTCAGTTCCGCGAGAATCTCCAACGTCACGAAAAGGAACTAGAACGAACCAGCCAGCAGATCAAGCGGCTCATCAAAGAAGTCAAGGATGTCGTGCAGGCAGCTAAACGTCTCGGCAGCGCTCAGCTGGCGCTGGCATCGAGCATGGAGCAGTTCGAGTTCGCCTGCATCGGTGCATCCATGACGGAAGACGAGCGCGTTATAGGACGCTCCCTGCACCACTTCGCCCATCTAATAAGGACCATAGAGGAAGAGAGGGAACGAATGTTGGGCAGAGCGCACGAGCAAATAATTCAGCCCCTGGAGAAGTTCCGGAAAGAACATATCGGTGCTGTCAAGGAAGGAAAAAAGAAATTCGATAAGAAGACTGCGAAGTTCTGTCAAACGCAAGAACGCACGTTGTCGCTGTCTTTGAAGAAACCAGAGACTGTCTTCCAAGAGGCTGATGCGGCAATGGACATGGCCGAGCGCGACTTCTGCCAGGCCTCCCTCGAGTACGTGTTCCAGCTGCAAGCCGTCCAGGAGAGGAAGAAGTTCGAGCTGGTGGAGACGCTGCTCGGCTTCGTCTTCGGCTGGTGGACCTTCCACCACACGGCGCACGACGTCCACGCCGACGCCGAGCCCAGGGTGCGCGATCTGCAACTCAGGATACAACGAACGAGGAGTAATTTCGAAGAGACAAGCAAACAGACTGAATCACTCATGAAGAAGATGATGGAAGTGCGGCAAATGTCCAAAGAAGAGGACGGAAGCGACGACGGCCCCGGGGGCGTGTCCAGAGCCGGGTACCTGTTCCTCATGGAGAAAAAGGCCTTCGGTACGACGACGTGGAGCAAGCAGTTCTGCACGTACGAGAAGAGATCCCGGATCCTGACCCTGACGCCGTACAATCAGATCAACGTCAAAACGAGCTGGTCTCCCGAGGCCGTGTGTCTGTGCGGCGCCGTGGCTTGCGGTGAGGCGGCGGAGCGGCGTTGGTGCTGGGAGGCAATCCCCGCAGACCGCGAGCGCAGCCCCCTCACGCTGCAGGCGCTCTCGCAGCGGGACCGCGCCGCCTGGCTCGCGGCCGTCGACGAGCCCCCCGCCGAGAGACACCCGCAGAAAGCATCTGTGAACAGTGAAGAGTTTGCGTTAGACGAAACGGGATTCGCCTTCGTGCGGAAGCTGATATCCGTGATAGAGTCGAGGGGGCTGGAGGAGCAGGGACTGTACAGGGTGGCGGGCGTGGCGTCCAAGGTGTCGCGGCTGGTGGCGTGCGGCGCGCCGGGCCGCCGCCTGCCGCCCGCGCTCCACGACCCGCTGGAGTGGGAGACCAAGACCCTGACCTCCGCGCTCAAGAGCTACCTGCGCGCGCTGCCCGACCCGCTGCTCACCCGCGCGCTGCACGACCGCTTCATCGCCGTCGTCAAGTGCGAGCGGCGGGCGGAGCGCGCGGCGGGCGTGTGCGAGCTGGTGCGCGCGCTGCCGCCGCACAACCGGGACATGCTGCTGCTGGTGCTGGCACACCTGCGGAAGGTAGCGGCGAAGAGCGATAAGAACCTAATGTCGGAGTCGAACCTGGCGGTCTGCTTCGGGCCCACGCTGCTGAGGCCGGAGCGTGAGACGGTGGCGTCCATACTGGAGCTCAAGTTCTACAACGTGCTGGTGGAGACCATGCTCGAGAACTACGACGAGATCTTCGAGGCCGGCCCCCCCGCCCCGCCCGCGCACAACGGCCTGCTCAGAACGTCGCCGACCGCGTTACCGACAGCGTCGAGGAATGATATCAGCTTGGTAGGCGCGGGCGGCATAGCGGCCGGCGTGGGGCCCGGCGCGCGATACTCGCCCCACCACCACCAGCTGCTGCAACACTTCACCCACCCGCACCCCCTCGCTCATGCGGGAGTGGGGCGCTCCAGCAGCAGCTCGGCCGAGTCGGTGTCCAGCGCGTCGCCCCCGCCCGGAGCCCCGGCCTCCGCCCTCCGCCCGCAACGCTGCACGTGA
Protein
MGVGLQPLEFTECLADSPQFRENLQRHEKELERTSQQIKRLIKEVKDVVQAAKRLGSAQLALASSMEQFEFACIGASMTEDERVIGRSLHHFAHLIRTIEEERERMLGRAHEQIIQPLEKFRKEHIGAVKEGKKKFDKKTAKFCQTQERTLSLSLKKPETVFQEADAAMDMAERDFCQASLEYVFQLQAVQERKKFELVETLLGFVFGWWTFHHTAHDVHADAEPRVRDLQLRIQRTRSNFEETSKQTESLMKKMMEVRQMSKEEDGSDDGPGGVSRAGYLFLMEKKAFGTTTWSKQFCTYEKRSRILTLTPYNQINVKTSWSPEAVCLCGAVACGEAAERRWCWEAIPADRERSPLTLQALSQRDRAAWLAAVDEPPAERHPQKASVNSEEFALDETGFAFVRKLISVIESRGLEEQGLYRVAGVASKVSRLVACGAPGRRLPPALHDPLEWETKTLTSALKSYLRALPDPLLTRALHDRFIAVVKCERRAERAAGVCELVRALPPHNRDMLLLVLAHLRKVAAKSDKNLMSESNLAVCFGPTLLRPERETVASILELKFYNVLVETMLENYDEIFEAGPPAPPAHNGLLRTSPTALPTASRNDISLVGAGGIAAGVGPGARYSPHHHQLLQHFTHPHPLAHAGVGRSSSSSAESVSSASPPPGAPASALRPQRCT
Summary
Uniprot
A0A194PY97
A0A212FC52
A0A191UR79
A0A0C9Q743
A0A154PE05
A0A0M9A447
+ More
A0A087ZRM9 A0A0L7R5T3 A0A1B6K0Y2 E2AEJ4 A0A1B6KCG6 A0A1B6KSM0 A0A1B6L4C8 A0A1B6L5F2 A0A232FDI1 A0A195BWM0 F4WRU0 A0A151XIT4 A0A158NEM0 A0A195CRI1 A0A195FKM3 A0A023F3M9 A0A067R0J4 A0A026WRR8 E2BT36 K7ITA3 A0A195ENT9 A0A0P5QT38 A0A3L8DEC5 A0A0P5I269 A0A0P5DKE9 A0A0P5GHV4 A0A0P5GNE1 A0A0P5GWH8 A0A0P5R2Y1 A0A0P5GMF9 A0A0P5I2A3 A0A0P5I814 A0A0P5HIV5 A0A0N8A1Q2 A0A0P5JZC5 A0A0P5HFQ8 A0A0P5Q2C3 A0A0P6IUU3 A0A0P6FRR2 A0A0P5B1D4 A0A0P5GBY7 A0A0P5HMV5 A0A0P5YUP9 A0A0P6AGY0 A0A0P5KVT1 A0A0P6GAC5 A0A1Y1LRG4 A0A1Y1LRF5 A0A2A3ER43 E9GSI6 A0A2H8TFJ3 A0A2S2NMQ4 J9JV82 T1HVB1 V5G502 A0A310SU94 A0A1S4EA17 A0A0A9ZDM2 A0A0K8T650 A0A0P5BCG3 A0A131Z5H7 A0A1S3JSA8 A0A0N8A5X1 A0A1Q3FTJ4 A0A1Q3FTK4 A0A131Z3Q1 A0A131YFE9 T1JFW8 A0A224Z5N1 A0A0P4YQN3 A0A182FLX3 A0A131XEE6 L7M711 A0A1E1X5U5 A0A2M4CHA7 A0A2M4CHL7 A0A0K8VHD9 A0A293LMB8 A0A0K2UZF5 A0A1S3JSF9 A0A0P5CD45 A0A0K8TZG8 A0A034VIS9 V5HZR0 A0A182QSY8 F5HLS9 A0A0P4YIE5 A0A1B0C9A1
A0A087ZRM9 A0A0L7R5T3 A0A1B6K0Y2 E2AEJ4 A0A1B6KCG6 A0A1B6KSM0 A0A1B6L4C8 A0A1B6L5F2 A0A232FDI1 A0A195BWM0 F4WRU0 A0A151XIT4 A0A158NEM0 A0A195CRI1 A0A195FKM3 A0A023F3M9 A0A067R0J4 A0A026WRR8 E2BT36 K7ITA3 A0A195ENT9 A0A0P5QT38 A0A3L8DEC5 A0A0P5I269 A0A0P5DKE9 A0A0P5GHV4 A0A0P5GNE1 A0A0P5GWH8 A0A0P5R2Y1 A0A0P5GMF9 A0A0P5I2A3 A0A0P5I814 A0A0P5HIV5 A0A0N8A1Q2 A0A0P5JZC5 A0A0P5HFQ8 A0A0P5Q2C3 A0A0P6IUU3 A0A0P6FRR2 A0A0P5B1D4 A0A0P5GBY7 A0A0P5HMV5 A0A0P5YUP9 A0A0P6AGY0 A0A0P5KVT1 A0A0P6GAC5 A0A1Y1LRG4 A0A1Y1LRF5 A0A2A3ER43 E9GSI6 A0A2H8TFJ3 A0A2S2NMQ4 J9JV82 T1HVB1 V5G502 A0A310SU94 A0A1S4EA17 A0A0A9ZDM2 A0A0K8T650 A0A0P5BCG3 A0A131Z5H7 A0A1S3JSA8 A0A0N8A5X1 A0A1Q3FTJ4 A0A1Q3FTK4 A0A131Z3Q1 A0A131YFE9 T1JFW8 A0A224Z5N1 A0A0P4YQN3 A0A182FLX3 A0A131XEE6 L7M711 A0A1E1X5U5 A0A2M4CHA7 A0A2M4CHL7 A0A0K8VHD9 A0A293LMB8 A0A0K2UZF5 A0A1S3JSF9 A0A0P5CD45 A0A0K8TZG8 A0A034VIS9 V5HZR0 A0A182QSY8 F5HLS9 A0A0P4YIE5 A0A1B0C9A1
Pubmed
EMBL
KQ459585
KPI98307.1
AGBW02009223
OWR51311.1
KU365968
ANJ04684.1
+ More
GBYB01009963 JAG79730.1 KQ434886 KZC10092.1 KQ435740 KOX76827.1 KQ414648 KOC66174.1 GECU01002601 JAT05106.1 GL438862 EFN68151.1 GEBQ01030834 JAT09143.1 GEBQ01025550 JAT14427.1 GEBQ01026999 GEBQ01021461 JAT12978.1 JAT18516.1 GEBQ01021116 GEBQ01001608 JAT18861.1 JAT38369.1 NNAY01000418 OXU28543.1 KQ976401 KYM92368.1 GL888292 EGI63097.1 KQ982080 KYQ60304.1 ADTU01013497 KQ977381 KYN03102.1 KQ981490 KYN41215.1 GBBI01002741 JAC15971.1 KK852794 KDR16415.1 KK107119 EZA58745.1 GL450325 EFN81166.1 AAZX01003313 AAZX01008304 KQ978625 KYN29577.1 GDIQ01110330 JAL41396.1 QOIP01000009 RLU18830.1 GDIQ01221591 JAK30134.1 GDIP01160970 JAJ62432.1 GDIQ01240439 JAK11286.1 GDIQ01241888 JAK09837.1 GDIQ01238734 JAK12991.1 GDIQ01112076 JAL39650.1 GDIQ01240438 JAK11287.1 GDIQ01219090 JAK32635.1 GDIQ01240437 JAK11288.1 GDIQ01232780 JAK18945.1 GDIP01191121 JAJ32281.1 GDIQ01193146 JAK58579.1 GDIQ01228582 JAK23143.1 GDIQ01120120 JAL31606.1 GDIQ01027449 JAN67288.1 GDIQ01061442 JAN33295.1 GDIP01191120 JAJ32282.1 GDIQ01243286 JAK08439.1 GDIQ01230836 JAK20889.1 GDIP01053154 JAM50561.1 GDIP01029396 JAM74319.1 GDIQ01178743 JAK72982.1 GDIQ01046605 JAN48132.1 GEZM01048972 GEZM01048971 JAV76233.1 GEZM01048973 JAV76232.1 KZ288202 PBC33511.1 GL732562 EFX77433.1 GFXV01000233 MBW12038.1 GGMR01005852 MBY18471.1 ABLF02040615 ABLF02040617 ACPB03009283 ACPB03009284 ACPB03009285 ACPB03009286 GALX01003336 JAB65130.1 KQ759874 OAD62359.1 GBHO01037387 GBHO01003689 GBHO01003688 GDHC01005368 JAG06217.1 JAG39915.1 JAG39916.1 JAQ13261.1 GBRD01005156 JAG60665.1 GDIP01201925 JAJ21477.1 GEDV01003186 JAP85371.1 GDIP01179369 JAJ44033.1 GFDL01004114 JAV30931.1 GFDL01004115 JAV30930.1 GEDV01003187 JAP85370.1 GEDV01011355 JAP77202.1 JH432190 GFPF01010354 MAA21500.1 GDIP01225274 JAI98127.1 GEFH01004580 JAP64001.1 GACK01005194 JAA59840.1 GFAC01004558 JAT94630.1 GGFL01000549 MBW64727.1 GGFL01000547 MBW64725.1 GDHF01014013 JAI38301.1 GFWV01009908 MAA34637.1 HACA01026079 CDW43440.1 GDIP01172335 JAJ51067.1 GDHF01032646 JAI19668.1 GAKP01016548 JAC42404.1 GANP01004230 JAB80238.1 AXCN02002217 AAAB01008949 EGK97240.1 GDIP01229523 JAI93878.1 AJWK01002077
GBYB01009963 JAG79730.1 KQ434886 KZC10092.1 KQ435740 KOX76827.1 KQ414648 KOC66174.1 GECU01002601 JAT05106.1 GL438862 EFN68151.1 GEBQ01030834 JAT09143.1 GEBQ01025550 JAT14427.1 GEBQ01026999 GEBQ01021461 JAT12978.1 JAT18516.1 GEBQ01021116 GEBQ01001608 JAT18861.1 JAT38369.1 NNAY01000418 OXU28543.1 KQ976401 KYM92368.1 GL888292 EGI63097.1 KQ982080 KYQ60304.1 ADTU01013497 KQ977381 KYN03102.1 KQ981490 KYN41215.1 GBBI01002741 JAC15971.1 KK852794 KDR16415.1 KK107119 EZA58745.1 GL450325 EFN81166.1 AAZX01003313 AAZX01008304 KQ978625 KYN29577.1 GDIQ01110330 JAL41396.1 QOIP01000009 RLU18830.1 GDIQ01221591 JAK30134.1 GDIP01160970 JAJ62432.1 GDIQ01240439 JAK11286.1 GDIQ01241888 JAK09837.1 GDIQ01238734 JAK12991.1 GDIQ01112076 JAL39650.1 GDIQ01240438 JAK11287.1 GDIQ01219090 JAK32635.1 GDIQ01240437 JAK11288.1 GDIQ01232780 JAK18945.1 GDIP01191121 JAJ32281.1 GDIQ01193146 JAK58579.1 GDIQ01228582 JAK23143.1 GDIQ01120120 JAL31606.1 GDIQ01027449 JAN67288.1 GDIQ01061442 JAN33295.1 GDIP01191120 JAJ32282.1 GDIQ01243286 JAK08439.1 GDIQ01230836 JAK20889.1 GDIP01053154 JAM50561.1 GDIP01029396 JAM74319.1 GDIQ01178743 JAK72982.1 GDIQ01046605 JAN48132.1 GEZM01048972 GEZM01048971 JAV76233.1 GEZM01048973 JAV76232.1 KZ288202 PBC33511.1 GL732562 EFX77433.1 GFXV01000233 MBW12038.1 GGMR01005852 MBY18471.1 ABLF02040615 ABLF02040617 ACPB03009283 ACPB03009284 ACPB03009285 ACPB03009286 GALX01003336 JAB65130.1 KQ759874 OAD62359.1 GBHO01037387 GBHO01003689 GBHO01003688 GDHC01005368 JAG06217.1 JAG39915.1 JAG39916.1 JAQ13261.1 GBRD01005156 JAG60665.1 GDIP01201925 JAJ21477.1 GEDV01003186 JAP85371.1 GDIP01179369 JAJ44033.1 GFDL01004114 JAV30931.1 GFDL01004115 JAV30930.1 GEDV01003187 JAP85370.1 GEDV01011355 JAP77202.1 JH432190 GFPF01010354 MAA21500.1 GDIP01225274 JAI98127.1 GEFH01004580 JAP64001.1 GACK01005194 JAA59840.1 GFAC01004558 JAT94630.1 GGFL01000549 MBW64727.1 GGFL01000547 MBW64725.1 GDHF01014013 JAI38301.1 GFWV01009908 MAA34637.1 HACA01026079 CDW43440.1 GDIP01172335 JAJ51067.1 GDHF01032646 JAI19668.1 GAKP01016548 JAC42404.1 GANP01004230 JAB80238.1 AXCN02002217 AAAB01008949 EGK97240.1 GDIP01229523 JAI93878.1 AJWK01002077
Proteomes
UP000053268
UP000007151
UP000076502
UP000053105
UP000005203
UP000053825
+ More
UP000000311 UP000215335 UP000078540 UP000007755 UP000075809 UP000005205 UP000078542 UP000078541 UP000027135 UP000053097 UP000008237 UP000002358 UP000078492 UP000279307 UP000242457 UP000000305 UP000007819 UP000015103 UP000079169 UP000085678 UP000069272 UP000075886 UP000007062 UP000092461
UP000000311 UP000215335 UP000078540 UP000007755 UP000075809 UP000005205 UP000078542 UP000078541 UP000027135 UP000053097 UP000008237 UP000002358 UP000078492 UP000279307 UP000242457 UP000000305 UP000007819 UP000015103 UP000079169 UP000085678 UP000069272 UP000075886 UP000007062 UP000092461
Interpro
Gene 3D
ProteinModelPortal
A0A194PY97
A0A212FC52
A0A191UR79
A0A0C9Q743
A0A154PE05
A0A0M9A447
+ More
A0A087ZRM9 A0A0L7R5T3 A0A1B6K0Y2 E2AEJ4 A0A1B6KCG6 A0A1B6KSM0 A0A1B6L4C8 A0A1B6L5F2 A0A232FDI1 A0A195BWM0 F4WRU0 A0A151XIT4 A0A158NEM0 A0A195CRI1 A0A195FKM3 A0A023F3M9 A0A067R0J4 A0A026WRR8 E2BT36 K7ITA3 A0A195ENT9 A0A0P5QT38 A0A3L8DEC5 A0A0P5I269 A0A0P5DKE9 A0A0P5GHV4 A0A0P5GNE1 A0A0P5GWH8 A0A0P5R2Y1 A0A0P5GMF9 A0A0P5I2A3 A0A0P5I814 A0A0P5HIV5 A0A0N8A1Q2 A0A0P5JZC5 A0A0P5HFQ8 A0A0P5Q2C3 A0A0P6IUU3 A0A0P6FRR2 A0A0P5B1D4 A0A0P5GBY7 A0A0P5HMV5 A0A0P5YUP9 A0A0P6AGY0 A0A0P5KVT1 A0A0P6GAC5 A0A1Y1LRG4 A0A1Y1LRF5 A0A2A3ER43 E9GSI6 A0A2H8TFJ3 A0A2S2NMQ4 J9JV82 T1HVB1 V5G502 A0A310SU94 A0A1S4EA17 A0A0A9ZDM2 A0A0K8T650 A0A0P5BCG3 A0A131Z5H7 A0A1S3JSA8 A0A0N8A5X1 A0A1Q3FTJ4 A0A1Q3FTK4 A0A131Z3Q1 A0A131YFE9 T1JFW8 A0A224Z5N1 A0A0P4YQN3 A0A182FLX3 A0A131XEE6 L7M711 A0A1E1X5U5 A0A2M4CHA7 A0A2M4CHL7 A0A0K8VHD9 A0A293LMB8 A0A0K2UZF5 A0A1S3JSF9 A0A0P5CD45 A0A0K8TZG8 A0A034VIS9 V5HZR0 A0A182QSY8 F5HLS9 A0A0P4YIE5 A0A1B0C9A1
A0A087ZRM9 A0A0L7R5T3 A0A1B6K0Y2 E2AEJ4 A0A1B6KCG6 A0A1B6KSM0 A0A1B6L4C8 A0A1B6L5F2 A0A232FDI1 A0A195BWM0 F4WRU0 A0A151XIT4 A0A158NEM0 A0A195CRI1 A0A195FKM3 A0A023F3M9 A0A067R0J4 A0A026WRR8 E2BT36 K7ITA3 A0A195ENT9 A0A0P5QT38 A0A3L8DEC5 A0A0P5I269 A0A0P5DKE9 A0A0P5GHV4 A0A0P5GNE1 A0A0P5GWH8 A0A0P5R2Y1 A0A0P5GMF9 A0A0P5I2A3 A0A0P5I814 A0A0P5HIV5 A0A0N8A1Q2 A0A0P5JZC5 A0A0P5HFQ8 A0A0P5Q2C3 A0A0P6IUU3 A0A0P6FRR2 A0A0P5B1D4 A0A0P5GBY7 A0A0P5HMV5 A0A0P5YUP9 A0A0P6AGY0 A0A0P5KVT1 A0A0P6GAC5 A0A1Y1LRG4 A0A1Y1LRF5 A0A2A3ER43 E9GSI6 A0A2H8TFJ3 A0A2S2NMQ4 J9JV82 T1HVB1 V5G502 A0A310SU94 A0A1S4EA17 A0A0A9ZDM2 A0A0K8T650 A0A0P5BCG3 A0A131Z5H7 A0A1S3JSA8 A0A0N8A5X1 A0A1Q3FTJ4 A0A1Q3FTK4 A0A131Z3Q1 A0A131YFE9 T1JFW8 A0A224Z5N1 A0A0P4YQN3 A0A182FLX3 A0A131XEE6 L7M711 A0A1E1X5U5 A0A2M4CHA7 A0A2M4CHL7 A0A0K8VHD9 A0A293LMB8 A0A0K2UZF5 A0A1S3JSF9 A0A0P5CD45 A0A0K8TZG8 A0A034VIS9 V5HZR0 A0A182QSY8 F5HLS9 A0A0P4YIE5 A0A1B0C9A1
PDB
1F7C
E-value=4.74724e-46,
Score=467
Ontologies
GO
Topology
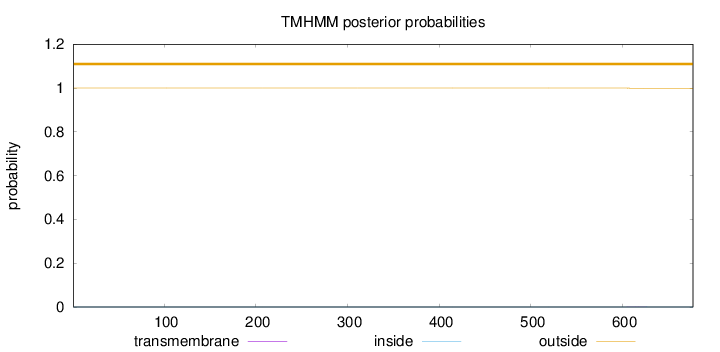
Length:
677
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 677
Population Genetic Test Statistics
Pi
221.654066
Theta
167.670581
Tajima's D
1.171225
CLR
0.373475
CSRT
0.705614719264037
Interpretation
Uncertain
