Pre Gene Modal
BGIBMGA009473
Annotation
PREDICTED:_apoptosis-inducing_factor_1?_mitochondrial-like_[Bombyx_mori]
Full name
Putative apoptosis-inducing factor 1, mitochondrial
Location in the cell
Mitochondrial Reliability : 1.398
Sequence
CDS
ATGCTCAGAGCGGTTTCCGTGTTGTGTGGAGAAAGTCAATCTTTTTTACTTTATACCACCGCTAGGAGAGATGCCTACAAACTCTTCCGTAATACATATGCCTCTGCCTCGACATCATCACAGACTGTCCTTACATCGGAAGCATCACAAGCATCACACAGCCCCAGTGTACATTCACCCCCTCCTGCACAATCAAGTCCACCTCCTACACCGCCATCACAGTCTCCGCGGCGATCATGGACTGTTGCAGCAGCCCTAGCACTCACCGCATCGGTTGGTGGAATATATTTCTTTCGTAAGCTGAGTGAAGAAGAAAAGAAAATCATATCCTCTGAAGATGCACCAAAAAAGAATGTTGGGCCAGTACGGACACCCATCCCGGCGTCTGCGGATCAGCTCCCGCGACACGTACAGTTCCTGCTGGTCGGTGCTGGTACTGCCTCGTTTGCAGCCATGAGGGCGATACGCAGTCAAAAACCCGACGCCCAGGTGCTGATGGTGGGTTCGGAGTACCAGCTGCCGTACATGCGGCCTCCGCTCACCAAGGAGCTGTGGCGGGAGCCAGACCTGGACAAAGTCACAGACCTCGACTCCTTGACCTTTAAACAGTGGAACGGCAGGGTGAGAAGCCTGGCGTACGAGCCGAGCGCGTTCTACACGCCCGTGGAGCAGCTGGAGGAGCGCGGCGCGGGGTGCGGGGTGGCGCGGGGCTGGGCGGTGCGGCGCCTGGACGTGGCGCGGCGCCGGGCCACGCTGGAGGGCGCGGGGGGCACGCACACGCTCACGTACGACTACTGCCTCGTCGCGACCGGCGTGGTGGCGCGGCGCTCGGCGGCGCTGGCGGGCGCGCGGGGCGCGGGGCGGGCGCGCACGCTGCGCACGGCGGATGACGTCACGCAGCTGGCGGCGCGGCTGGCGGGGGGGGACGTGCAGCGCGTGGCGGTGCTGGGCGGGGGGCTGCTGGCCACCGAGCTCAGCGCCGCGCTCGCCGCCAGATTAAAGGATACGGGTAAAACAGTATTAGAATTGTACCGAGAGAGTAGTCCTCTAGCGTCCATCCTGCCCTCCTACCTCGCGGAGGAGGCCGCCCGGAGGCTCCAAGAAGCGGGGGTCACACTCCTGCCTGAAGCGGACGTGGTAGATTCTATAGTGGTTGATGACAAAGTGCTGATGCGACTTGCGAGTGGCGCCGAGGTGGAGGCCGACTACGTGGTGGAGTGTGTCGGCACGGAGTTCGATCCCGCGCTGGCCGACGCCTCCGAGCTGGAGGTTCACCCCGAGCTGGGCGGGGTGGTCGTCAACCCTGAGCTGCAGGCCCCAACATAA
Protein
MLRAVSVLCGESQSFLLYTTARRDAYKLFRNTYASASTSSQTVLTSEASQASHSPSVHSPPPAQSSPPPTPPSQSPRRSWTVAAALALTASVGGIYFFRKLSEEEKKIISSEDAPKKNVGPVRTPIPASADQLPRHVQFLLVGAGTASFAAMRAIRSQKPDAQVLMVGSEYQLPYMRPPLTKELWREPDLDKVTDLDSLTFKQWNGRVRSLAYEPSAFYTPVEQLEERGAGCGVARGWAVRRLDVARRRATLEGAGGTHTLTYDYCLVATGVVARRSAALAGARGAGRARTLRTADDVTQLAARLAGGDVQRVAVLGGGLLATELSAALAARLKDTGKTVLELYRESSPLASILPSYLAEEAARRLQEAGVTLLPEADVVDSIVVDDKVLMRLASGAEVEADYVVECVGTEFDPALADASELEVHPELGGVVVNPELQAPT
Summary
Description
Probable NADH oxidoreductase (By similarity). Mitochondrial effector of cell death that plays roles in developmentally regulated cell death and normal mitochondrial function.
Cofactor
FAD
Similarity
Belongs to the FAD-dependent oxidoreductase family.
Keywords
Alternative splicing
Apoptosis
Complete proteome
FAD
Flavoprotein
Mitochondrion
NAD
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Putative apoptosis-inducing factor 1, mitochondrial
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A2H1W831
A0A212FC61
A0A0N1PH97
A0A3S2LV53
A0A194PZY4
A0A0C9QS22
+ More
A0A1B6DIK6 A0A1Y1LC16 B4G966 Q29MI5 A0A0R3NY98 A0A0Q9X3C3 B4KGB9 B4JAR0 A0A151K0C0 A0A0K8VG98 A0A0K8U3V8 A0A0K8WFS2 A0A034WSC9 A0A2H8TVU6 A0A0P5Q211 B4MVU2 V5GXJ4 A0A0K8URX9 A0A034WNU7 A0A0K8V3Q0 A0A034WSR9 A0A0P5FRE2 A0A0P5VK51 A0A2S2PEQ0 A0A0P4WP23 A0A0P5BQF3 A0A151IAI0 A0A0P5Q6U6 A0A0P5I6F7 A0A0P5HWQ5 B4Q7N0 A0A0P5SLX2 A0A0P5T8B7 A0A0P5BBX7 A0A0J9QU21 A0A0P6GCM6 A0A0N8EEA1 A0A0P5RV82 A0A0P5RU69 A0A0P4Y062 A0A0P5J0M1 A0A0P5JEU1 A0A0P4WPP2 A0A0P4WSA2 A0A0P6G8K0 A0A0P5ATI5 A0A0P5BJ72 A0A0P5AQW0 A0A0P5XIR9 A0A0P5BIH7 A0A0P6FP79 A0A0P5H8Z7 A0A0P6B0G9 A0A0P5E3W8 A0A0P5YAJ9 A0A0N8C166 A0A151J8E8 A0A0P5QDI6 A0A0P4ZUI1 A0A0P4YA23 A0A0N7ZEH1 A0A0P5WI87 A0A0P5KP06 A0A0P5ECD8 J9JWN7 A0A0P5ZZC3 A0A0P5Q058 A0A0P5EKU5 A0A0N8EBC1 Q9VQ79-1 A0A0P5XGU3 B4IM59 A0A0N8E409 A0A0R1DL02 A0A0P5J329 A0A0P5GR20 A0A0P5EVM0 A0A0P5PJB6 A0A0P6FLN1 A0A0P4WTB0 A0A0N8ARE7 A0A0P4Z9V0 A0A0Q5WI30 A0A0P5IFX4 A0A0P6GHA3 A0A0P5HGL4 A0A0P5WIY5 A0A0N8E8W3 A0A0J7L085 A0A0P5SYR0 A0A0P4Z3H5 A0A0P5GMA5
A0A1B6DIK6 A0A1Y1LC16 B4G966 Q29MI5 A0A0R3NY98 A0A0Q9X3C3 B4KGB9 B4JAR0 A0A151K0C0 A0A0K8VG98 A0A0K8U3V8 A0A0K8WFS2 A0A034WSC9 A0A2H8TVU6 A0A0P5Q211 B4MVU2 V5GXJ4 A0A0K8URX9 A0A034WNU7 A0A0K8V3Q0 A0A034WSR9 A0A0P5FRE2 A0A0P5VK51 A0A2S2PEQ0 A0A0P4WP23 A0A0P5BQF3 A0A151IAI0 A0A0P5Q6U6 A0A0P5I6F7 A0A0P5HWQ5 B4Q7N0 A0A0P5SLX2 A0A0P5T8B7 A0A0P5BBX7 A0A0J9QU21 A0A0P6GCM6 A0A0N8EEA1 A0A0P5RV82 A0A0P5RU69 A0A0P4Y062 A0A0P5J0M1 A0A0P5JEU1 A0A0P4WPP2 A0A0P4WSA2 A0A0P6G8K0 A0A0P5ATI5 A0A0P5BJ72 A0A0P5AQW0 A0A0P5XIR9 A0A0P5BIH7 A0A0P6FP79 A0A0P5H8Z7 A0A0P6B0G9 A0A0P5E3W8 A0A0P5YAJ9 A0A0N8C166 A0A151J8E8 A0A0P5QDI6 A0A0P4ZUI1 A0A0P4YA23 A0A0N7ZEH1 A0A0P5WI87 A0A0P5KP06 A0A0P5ECD8 J9JWN7 A0A0P5ZZC3 A0A0P5Q058 A0A0P5EKU5 A0A0N8EBC1 Q9VQ79-1 A0A0P5XGU3 B4IM59 A0A0N8E409 A0A0R1DL02 A0A0P5J329 A0A0P5GR20 A0A0P5EVM0 A0A0P5PJB6 A0A0P6FLN1 A0A0P4WTB0 A0A0N8ARE7 A0A0P4Z9V0 A0A0Q5WI30 A0A0P5IFX4 A0A0P6GHA3 A0A0P5HGL4 A0A0P5WIY5 A0A0N8E8W3 A0A0J7L085 A0A0P5SYR0 A0A0P4Z3H5 A0A0P5GMA5
EC Number
1.1.1.-
Pubmed
EMBL
ODYU01006927
SOQ49207.1
AGBW02009223
OWR51313.1
KQ460889
KPJ11067.1
+ More
RSAL01000178 RVE45002.1 KQ459585 KPI98304.1 GBYB01003442 JAG73209.1 GEDC01011769 JAS25529.1 GEZM01061486 JAV70438.1 CH479180 EDW28896.1 CH379060 EAL33709.3 KRT04133.1 CH933807 KRG02527.1 EDW11106.2 CH916368 EDW03868.1 KQ981319 KYN42729.1 GDHF01029919 GDHF01014446 GDHF01007738 GDHF01004258 JAI22395.1 JAI37868.1 JAI44576.1 JAI48056.1 GDHF01030900 JAI21414.1 GDHF01002298 JAI50016.1 GAKP01001750 JAC57202.1 GFXV01006234 MBW18039.1 GDIQ01120229 JAL31497.1 CH963857 EDW75812.2 GALX01001969 JAB66497.1 GDHF01022862 JAI29452.1 GAKP01001751 JAC57201.1 GDHF01018886 JAI33428.1 GAKP01001752 JAC57200.1 GDIQ01254295 JAJ97429.1 GDIP01100426 JAM03289.1 GGMR01014767 MBY27386.1 GDIP01252765 JAI70636.1 GDIP01185335 JAJ38067.1 KQ978221 KYM96276.1 GDIQ01124930 JAL26796.1 GDIQ01218781 JAK32944.1 GDIQ01221184 JAK30541.1 CM000361 CM002910 EDX03410.1 KMY87587.1 GDIP01138138 JAL65576.1 GDIP01129578 JAL74136.1 GDIP01186631 JAJ36771.1 KMY87588.1 GDIQ01035261 JAN59476.1 GDIQ01035260 JAN59477.1 GDIQ01096183 JAL55543.1 GDIQ01096184 JAL55542.1 GDIP01234431 JAI88970.1 GDIQ01204837 JAK46888.1 GDIQ01206351 JAK45374.1 GDIP01253412 JAI69989.1 GDIP01253370 JAI70031.1 GDIQ01036970 JAN57767.1 GDIP01199071 JAJ24331.1 GDIP01184131 JAJ39271.1 GDIP01209724 JAJ13678.1 GDIP01071501 JAM32214.1 GDIP01199072 JAJ24330.1 GDIQ01047456 JAN47281.1 GDIQ01236431 JAK15294.1 GDIP01035702 JAM68013.1 GDIP01196886 GDIP01148483 LRGB01001361 JAJ74919.1 KZS12716.1 GDIP01061989 JAM41726.1 GDIQ01124931 JAL26795.1 KQ979533 KYN21118.1 GDIQ01115631 GDIQ01115630 JAL36096.1 GDIP01208533 JAJ14869.1 GDIP01230526 JAI92875.1 GDIP01253369 JAI70032.1 GDIP01086083 JAM17632.1 GDIQ01181298 JAK70427.1 GDIP01148484 JAJ74918.1 ABLF02015073 GDIP01037003 JAM66712.1 GDIQ01128124 JAL23602.1 GDIQ01270706 JAJ81018.1 GDIQ01043500 JAN51237.1 AE014134 AY052083 AY058696 BT044192 GDIP01084635 JAM19080.1 CH480936 EDW44539.1 GDIQ01063996 JAN30741.1 CM000157 KRJ97117.1 GDIQ01251733 GDIQ01216594 GDIQ01207701 JAK44024.1 GDIQ01238838 JAK12887.1 GDIQ01267305 JAJ84419.1 GDIQ01137828 JAL13898.1 GDIQ01045657 JAN49080.1 GDIP01251116 JAI72285.1 GDIQ01250283 JAK01442.1 GDIP01218981 JAJ04421.1 CH954177 KQS69982.1 GDIQ01217685 JAK34040.1 GDIQ01045658 JAN49079.1 GDIQ01250284 JAK01441.1 GDIP01086082 JAM17633.1 GDIQ01050364 JAN44373.1 LBMM01001670 KMQ95914.1 GDIP01138137 JAL65577.1 GDIP01218982 JAJ04420.1 GDIQ01245846 GDIQ01245845 JAK05880.1
RSAL01000178 RVE45002.1 KQ459585 KPI98304.1 GBYB01003442 JAG73209.1 GEDC01011769 JAS25529.1 GEZM01061486 JAV70438.1 CH479180 EDW28896.1 CH379060 EAL33709.3 KRT04133.1 CH933807 KRG02527.1 EDW11106.2 CH916368 EDW03868.1 KQ981319 KYN42729.1 GDHF01029919 GDHF01014446 GDHF01007738 GDHF01004258 JAI22395.1 JAI37868.1 JAI44576.1 JAI48056.1 GDHF01030900 JAI21414.1 GDHF01002298 JAI50016.1 GAKP01001750 JAC57202.1 GFXV01006234 MBW18039.1 GDIQ01120229 JAL31497.1 CH963857 EDW75812.2 GALX01001969 JAB66497.1 GDHF01022862 JAI29452.1 GAKP01001751 JAC57201.1 GDHF01018886 JAI33428.1 GAKP01001752 JAC57200.1 GDIQ01254295 JAJ97429.1 GDIP01100426 JAM03289.1 GGMR01014767 MBY27386.1 GDIP01252765 JAI70636.1 GDIP01185335 JAJ38067.1 KQ978221 KYM96276.1 GDIQ01124930 JAL26796.1 GDIQ01218781 JAK32944.1 GDIQ01221184 JAK30541.1 CM000361 CM002910 EDX03410.1 KMY87587.1 GDIP01138138 JAL65576.1 GDIP01129578 JAL74136.1 GDIP01186631 JAJ36771.1 KMY87588.1 GDIQ01035261 JAN59476.1 GDIQ01035260 JAN59477.1 GDIQ01096183 JAL55543.1 GDIQ01096184 JAL55542.1 GDIP01234431 JAI88970.1 GDIQ01204837 JAK46888.1 GDIQ01206351 JAK45374.1 GDIP01253412 JAI69989.1 GDIP01253370 JAI70031.1 GDIQ01036970 JAN57767.1 GDIP01199071 JAJ24331.1 GDIP01184131 JAJ39271.1 GDIP01209724 JAJ13678.1 GDIP01071501 JAM32214.1 GDIP01199072 JAJ24330.1 GDIQ01047456 JAN47281.1 GDIQ01236431 JAK15294.1 GDIP01035702 JAM68013.1 GDIP01196886 GDIP01148483 LRGB01001361 JAJ74919.1 KZS12716.1 GDIP01061989 JAM41726.1 GDIQ01124931 JAL26795.1 KQ979533 KYN21118.1 GDIQ01115631 GDIQ01115630 JAL36096.1 GDIP01208533 JAJ14869.1 GDIP01230526 JAI92875.1 GDIP01253369 JAI70032.1 GDIP01086083 JAM17632.1 GDIQ01181298 JAK70427.1 GDIP01148484 JAJ74918.1 ABLF02015073 GDIP01037003 JAM66712.1 GDIQ01128124 JAL23602.1 GDIQ01270706 JAJ81018.1 GDIQ01043500 JAN51237.1 AE014134 AY052083 AY058696 BT044192 GDIP01084635 JAM19080.1 CH480936 EDW44539.1 GDIQ01063996 JAN30741.1 CM000157 KRJ97117.1 GDIQ01251733 GDIQ01216594 GDIQ01207701 JAK44024.1 GDIQ01238838 JAK12887.1 GDIQ01267305 JAJ84419.1 GDIQ01137828 JAL13898.1 GDIQ01045657 JAN49080.1 GDIP01251116 JAI72285.1 GDIQ01250283 JAK01442.1 GDIP01218981 JAJ04421.1 CH954177 KQS69982.1 GDIQ01217685 JAK34040.1 GDIQ01045658 JAN49079.1 GDIQ01250284 JAK01441.1 GDIP01086082 JAM17633.1 GDIQ01050364 JAN44373.1 LBMM01001670 KMQ95914.1 GDIP01138137 JAL65577.1 GDIP01218982 JAJ04420.1 GDIQ01245846 GDIQ01245845 JAK05880.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2H1W831
A0A212FC61
A0A0N1PH97
A0A3S2LV53
A0A194PZY4
A0A0C9QS22
+ More
A0A1B6DIK6 A0A1Y1LC16 B4G966 Q29MI5 A0A0R3NY98 A0A0Q9X3C3 B4KGB9 B4JAR0 A0A151K0C0 A0A0K8VG98 A0A0K8U3V8 A0A0K8WFS2 A0A034WSC9 A0A2H8TVU6 A0A0P5Q211 B4MVU2 V5GXJ4 A0A0K8URX9 A0A034WNU7 A0A0K8V3Q0 A0A034WSR9 A0A0P5FRE2 A0A0P5VK51 A0A2S2PEQ0 A0A0P4WP23 A0A0P5BQF3 A0A151IAI0 A0A0P5Q6U6 A0A0P5I6F7 A0A0P5HWQ5 B4Q7N0 A0A0P5SLX2 A0A0P5T8B7 A0A0P5BBX7 A0A0J9QU21 A0A0P6GCM6 A0A0N8EEA1 A0A0P5RV82 A0A0P5RU69 A0A0P4Y062 A0A0P5J0M1 A0A0P5JEU1 A0A0P4WPP2 A0A0P4WSA2 A0A0P6G8K0 A0A0P5ATI5 A0A0P5BJ72 A0A0P5AQW0 A0A0P5XIR9 A0A0P5BIH7 A0A0P6FP79 A0A0P5H8Z7 A0A0P6B0G9 A0A0P5E3W8 A0A0P5YAJ9 A0A0N8C166 A0A151J8E8 A0A0P5QDI6 A0A0P4ZUI1 A0A0P4YA23 A0A0N7ZEH1 A0A0P5WI87 A0A0P5KP06 A0A0P5ECD8 J9JWN7 A0A0P5ZZC3 A0A0P5Q058 A0A0P5EKU5 A0A0N8EBC1 Q9VQ79-1 A0A0P5XGU3 B4IM59 A0A0N8E409 A0A0R1DL02 A0A0P5J329 A0A0P5GR20 A0A0P5EVM0 A0A0P5PJB6 A0A0P6FLN1 A0A0P4WTB0 A0A0N8ARE7 A0A0P4Z9V0 A0A0Q5WI30 A0A0P5IFX4 A0A0P6GHA3 A0A0P5HGL4 A0A0P5WIY5 A0A0N8E8W3 A0A0J7L085 A0A0P5SYR0 A0A0P4Z3H5 A0A0P5GMA5
A0A1B6DIK6 A0A1Y1LC16 B4G966 Q29MI5 A0A0R3NY98 A0A0Q9X3C3 B4KGB9 B4JAR0 A0A151K0C0 A0A0K8VG98 A0A0K8U3V8 A0A0K8WFS2 A0A034WSC9 A0A2H8TVU6 A0A0P5Q211 B4MVU2 V5GXJ4 A0A0K8URX9 A0A034WNU7 A0A0K8V3Q0 A0A034WSR9 A0A0P5FRE2 A0A0P5VK51 A0A2S2PEQ0 A0A0P4WP23 A0A0P5BQF3 A0A151IAI0 A0A0P5Q6U6 A0A0P5I6F7 A0A0P5HWQ5 B4Q7N0 A0A0P5SLX2 A0A0P5T8B7 A0A0P5BBX7 A0A0J9QU21 A0A0P6GCM6 A0A0N8EEA1 A0A0P5RV82 A0A0P5RU69 A0A0P4Y062 A0A0P5J0M1 A0A0P5JEU1 A0A0P4WPP2 A0A0P4WSA2 A0A0P6G8K0 A0A0P5ATI5 A0A0P5BJ72 A0A0P5AQW0 A0A0P5XIR9 A0A0P5BIH7 A0A0P6FP79 A0A0P5H8Z7 A0A0P6B0G9 A0A0P5E3W8 A0A0P5YAJ9 A0A0N8C166 A0A151J8E8 A0A0P5QDI6 A0A0P4ZUI1 A0A0P4YA23 A0A0N7ZEH1 A0A0P5WI87 A0A0P5KP06 A0A0P5ECD8 J9JWN7 A0A0P5ZZC3 A0A0P5Q058 A0A0P5EKU5 A0A0N8EBC1 Q9VQ79-1 A0A0P5XGU3 B4IM59 A0A0N8E409 A0A0R1DL02 A0A0P5J329 A0A0P5GR20 A0A0P5EVM0 A0A0P5PJB6 A0A0P6FLN1 A0A0P4WTB0 A0A0N8ARE7 A0A0P4Z9V0 A0A0Q5WI30 A0A0P5IFX4 A0A0P6GHA3 A0A0P5HGL4 A0A0P5WIY5 A0A0N8E8W3 A0A0J7L085 A0A0P5SYR0 A0A0P4Z3H5 A0A0P5GMA5
PDB
5FS7
E-value=6.10846e-30,
Score=327
Ontologies
GO
Topology
Subcellular location
Mitochondrion intermembrane space
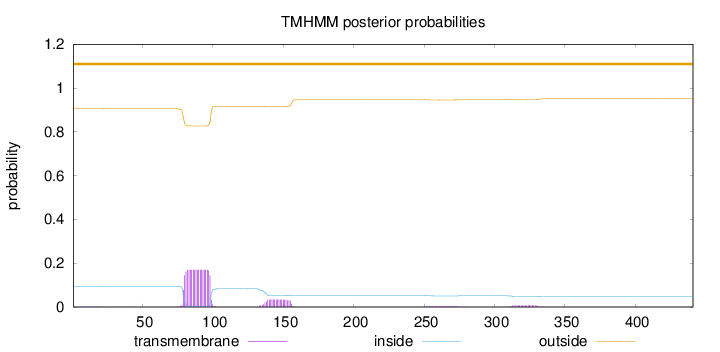
Length:
441
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.2041
Exp number, first 60 AAs:
0.0265
Total prob of N-in:
0.09354
outside
1 - 441
Population Genetic Test Statistics
Pi
180.230358
Theta
169.526299
Tajima's D
0.797057
CLR
0.784156
CSRT
0.602769861506925
Interpretation
Uncertain
