Gene
KWMTBOMO08187
Pre Gene Modal
BGIBMGA009488
Annotation
PREDICTED:_venom_acid_phosphatase_Acph-1-like_[Papilio_xuthus]
Full name
Prostatic acid phosphatase
Location in the cell
Cytoplasmic Reliability : 2.121
Sequence
CDS
ATGCTGAAATATTTAATATTCGTCTATTTGCTGATGCAAGTGAACACATCCCCCGGCAACGTCACCAACCGGGAGGCGAACGATGACATGGAACTGTTGATGGTTTATTTTGTGAACAGACATGGCGAGAGAGCTCCCGATGATGTGGAGCTTTCTCTTTCTGATCAGCAGGAAGAACTGAAGAGCCTCACTCACGTAGAAGGTCCTGAAGGCTTGACCAACGTCGGTAAAAGAAGGGCATACCAAATTGGCAAGTTCATCCGTCAGCGCTACGGATCCGAGGGTTCGAAAATCTTATCCAAAGTCTACCTGAAGGATGAGATTCTCGTTAGAAGCACAGATAAGGATCGGACGAAGATGACGGCGCTAGTAGCCATGTCTGCTGCCTACCCCCCGGAACCAGTTCAGCAATGGGATGAGTCACTGGGAAAGATCTGGCAGCCAGTCCCTTACTACTCTGTTCCACTAAACGAGGATTATCTTCGCTACTTCTCCAACTGCGACAGGTTTGTGGAGCTGATGAGGAACGCCAAGGACGAATCCATTCGGCAAGAATTCGCACCGTACCTGGACGTCATGCAGTTGCTCTCGAGGAGAACCGGCCAGAATTTTGTTGAGAACCCGCTGCTTCTGCAAGCCCTGTTCGACTTATTCAGGAGCTCGTTGGCCCTTGGTTTGAACATACCGGAGTGGGCAAAGCCCTTTCTGGAGAGCTTAAGCGAAGGAGCCAGGCTCGCCTACAAGTTGTACTTCAGAACTAATGAAATGAAAAAAATTGCTGGAGGGGTCCTACTCAACAAGTTCACAGAACAGGCCCTGATCGCGGCATCAGGTAAAGAGATCAAGCCCCGTCTCCACGTATTCTCCTCGCACGACTTCAACTTGGGCGCGTTCATGGAGGTGTCCAGGGTCCGCAGCAACCACGGCATCCCGGAGTACGGCGCTGCTTTCGCATTAGAACTGTACCGGTCGAAGGTCACAGGAGAACTTAGCGTCTTGCCGGTGTACCTTCCGCAAGCAGGAGACTCCACGCCCCAGGTGTTGCAGGTCAACGGCTGCGACGCAAACCCCTGCGGCCTGACAAAGTTCATGGAATTGACTAAAGATTTCCTACTACCCGAACATGAATATTACAGAATATGTAACATTAAGACAGAGTTATAA
Protein
MLKYLIFVYLLMQVNTSPGNVTNREANDDMELLMVYFVNRHGERAPDDVELSLSDQQEELKSLTHVEGPEGLTNVGKRRAYQIGKFIRQRYGSEGSKILSKVYLKDEILVRSTDKDRTKMTALVAMSAAYPPEPVQQWDESLGKIWQPVPYYSVPLNEDYLRYFSNCDRFVELMRNAKDESIRQEFAPYLDVMQLLSRRTGQNFVENPLLLQALFDLFRSSLALGLNIPEWAKPFLESLSEGARLAYKLYFRTNEMKKIAGGVLLNKFTEQALIAASGKEIKPRLHVFSSHDFNLGAFMEVSRVRSNHGIPEYGAAFALELYRSKVTGELSVLPVYLPQAGDSTPQVLQVNGCDANPCGLTKFMELTKDFLLPEHEYYRICNIKTEL
Summary
Description
A non-specific tyrosine phosphatase that dephosphorylates a diverse number of substrates under acidic conditions (pH 4-6) including alkyl, aryl, and acyl orthophosphate monoesters and phosphorylated proteins. Has lipid phosphatase activity and inactivates lysophosphatidic acid in seminal plasma (By similarity).
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Subunit
Homodimer; dimer formation is required for phosphatase activity.
Similarity
Belongs to the histidine acid phosphatase family.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Reference proteome
Secreted
Signal
Feature
chain Prostatic acid phosphatase
Uniprot
H9JIY7
A0A2A4JFS4
A0A437AY25
I4DR67
A0A1E1WMB7
A0A2H1VAX3
+ More
S4NSI3 A0A212EPG3 A0A194PY67 A0A0N0PBK2 A0A1E1W159 A0A0L7LBD9 H9J9I3 A0A2A4K986 A0A194PS41 A0A2H1W2H8 A0A3S2LGG3 A0A194R7T8 H9JXV1 A0A437B8H1 A0A212F630 A0A2H1WGF6 A0A2H1W2M4 I4DJY6 A0A2A4K8E9 A0A194PY59 A0A2A4K851 A0A2A4K724 A0A2S2NL16 K7IZV4 A0A1A9WGA5 A0A0L7LP95 A0A0C9RY44 J9K7T8 A0A1W6EVT0 A0A2H8TD02 C1IC99 A0A2U4B980 A0A2S2QXE2 J9K623 D3TNZ8 A0A0L7QWI3 A0A1A9ZSA2 A0A1B0BCK4 A0A1Q3EW11 A0A1Q3EVW8 A0A1Q3EW78 A0A1Q3EVW7 K7IZV5 A0A384A0X1 K7IQ63 Q17L85 K7IQ61 A0A340WSB5 K7IQ62 E2C1V8 A0A0M4EIW0 A0A1B0GD38 A0A3B0JV23 B4MXK6 A0A0K8TLP0 A0A369SGB3 F1SPF0 B0XEX3 A0A287AFE9 A0A1J1I6C6 A0A452EWS2 A0A232ESQ6 A0A341CZH3 A0A2A4K8T7 A0A1Y1KWW5 A0A195E0B2 L8IIS0 A0A1Y1KWT0 W5PH63 A6H730 A0A3Q1M6R0 A0A232EEI8 A0A023EQR2 A0A1L8DYV9 G3STE9 A0A210R3A2 A0A310SN00 B4H982 B4J1A8 F1Q4D3 A0A151P351 J3JY56 E2C1V9 A0A3Q7UDM5 A0A1W7R8D5 E2ARU9 A0A1W4WJR1 K9IJ78 A0A060GL20 A0A067QNZ9 F4WFK0
S4NSI3 A0A212EPG3 A0A194PY67 A0A0N0PBK2 A0A1E1W159 A0A0L7LBD9 H9J9I3 A0A2A4K986 A0A194PS41 A0A2H1W2H8 A0A3S2LGG3 A0A194R7T8 H9JXV1 A0A437B8H1 A0A212F630 A0A2H1WGF6 A0A2H1W2M4 I4DJY6 A0A2A4K8E9 A0A194PY59 A0A2A4K851 A0A2A4K724 A0A2S2NL16 K7IZV4 A0A1A9WGA5 A0A0L7LP95 A0A0C9RY44 J9K7T8 A0A1W6EVT0 A0A2H8TD02 C1IC99 A0A2U4B980 A0A2S2QXE2 J9K623 D3TNZ8 A0A0L7QWI3 A0A1A9ZSA2 A0A1B0BCK4 A0A1Q3EW11 A0A1Q3EVW8 A0A1Q3EW78 A0A1Q3EVW7 K7IZV5 A0A384A0X1 K7IQ63 Q17L85 K7IQ61 A0A340WSB5 K7IQ62 E2C1V8 A0A0M4EIW0 A0A1B0GD38 A0A3B0JV23 B4MXK6 A0A0K8TLP0 A0A369SGB3 F1SPF0 B0XEX3 A0A287AFE9 A0A1J1I6C6 A0A452EWS2 A0A232ESQ6 A0A341CZH3 A0A2A4K8T7 A0A1Y1KWW5 A0A195E0B2 L8IIS0 A0A1Y1KWT0 W5PH63 A6H730 A0A3Q1M6R0 A0A232EEI8 A0A023EQR2 A0A1L8DYV9 G3STE9 A0A210R3A2 A0A310SN00 B4H982 B4J1A8 F1Q4D3 A0A151P351 J3JY56 E2C1V9 A0A3Q7UDM5 A0A1W7R8D5 E2ARU9 A0A1W4WJR1 K9IJ78 A0A060GL20 A0A067QNZ9 F4WFK0
EC Number
3.1.3.2
Pubmed
EMBL
BABH01022999
BABH01023000
NWSH01001685
PCG70424.1
RSAL01000278
RVE43078.1
+ More
AK404965 BAM20407.1 GDQN01003058 JAT87996.1 ODYU01001406 SOQ37542.1 GAIX01014082 JAA78478.1 AGBW02013469 OWR43369.1 KQ459585 KPI98277.1 KQ460889 KPJ11041.1 GDQN01010326 JAT80728.1 JTDY01001815 KOB72803.1 BABH01004863 NWSH01000024 PCG80629.1 KQ459601 KPI93945.1 ODYU01005909 SOQ47289.1 RSAL01000014 RVE53289.1 KQ460855 KPJ11936.1 BABH01040368 RSAL01000117 RVE46811.1 AGBW02010086 OWR49186.1 ODYU01008500 SOQ52158.1 SOQ47287.1 AK401604 BAM18226.1 NWSH01000059 PCG80073.1 KPI98276.1 PCG80074.1 PCG80075.1 GGMR01005235 MBY17854.1 JTDY01000422 KOB77267.1 GBYB01012831 JAG82598.1 ABLF02038965 KY563421 ARK19830.1 GFXV01000178 MBW11983.1 EU365869 ACA60733.1 GGMS01013238 MBY82441.1 ABLF02019532 EZ423150 ADD19426.1 KQ414714 KOC62926.1 JXJN01012024 GFDL01015589 JAV19456.1 GFDL01015590 JAV19455.1 GFDL01015587 JAV19458.1 GFDL01015588 JAV19457.1 CH477217 EAT47490.1 GL452008 EFN78070.1 CP012525 ALC44752.1 CCAG010002313 CCAG010002314 OUUW01000002 SPP76591.1 CH963876 EDW76775.2 GDAI01002763 JAI14840.1 NOWV01000013 RDD45829.1 AEMK02000088 DS232876 EDS26304.1 CVRI01000042 CRK95747.1 LWLT01000001 NNAY01002389 OXU21381.1 PCG80631.1 GEZM01071381 GEZM01071374 JAV65852.1 KQ979955 KYN18558.1 JH881123 ELR56450.1 GEZM01071378 GEZM01071370 JAV65859.1 AMGL01011780 AMGL01011781 AMGL01011782 BC146093 NNAY01005308 OXU16770.1 GAPW01001661 JAC11937.1 GFDF01002423 JAV11661.1 NEDP02000657 OWF55488.1 KQ762196 OAD56135.1 CH479227 EDW35306.1 CH916366 EDV97977.1 AAEX03013595 AKHW03001210 KYO43349.1 BT128180 AEE63141.1 EFN78071.1 GEHC01000248 JAV47397.1 GL442175 EFN63832.1 GABZ01006540 JAA46985.1 KJ710422 AIB53032.1 KK853123 KDR11063.1 GL888120 EGI66977.1
AK404965 BAM20407.1 GDQN01003058 JAT87996.1 ODYU01001406 SOQ37542.1 GAIX01014082 JAA78478.1 AGBW02013469 OWR43369.1 KQ459585 KPI98277.1 KQ460889 KPJ11041.1 GDQN01010326 JAT80728.1 JTDY01001815 KOB72803.1 BABH01004863 NWSH01000024 PCG80629.1 KQ459601 KPI93945.1 ODYU01005909 SOQ47289.1 RSAL01000014 RVE53289.1 KQ460855 KPJ11936.1 BABH01040368 RSAL01000117 RVE46811.1 AGBW02010086 OWR49186.1 ODYU01008500 SOQ52158.1 SOQ47287.1 AK401604 BAM18226.1 NWSH01000059 PCG80073.1 KPI98276.1 PCG80074.1 PCG80075.1 GGMR01005235 MBY17854.1 JTDY01000422 KOB77267.1 GBYB01012831 JAG82598.1 ABLF02038965 KY563421 ARK19830.1 GFXV01000178 MBW11983.1 EU365869 ACA60733.1 GGMS01013238 MBY82441.1 ABLF02019532 EZ423150 ADD19426.1 KQ414714 KOC62926.1 JXJN01012024 GFDL01015589 JAV19456.1 GFDL01015590 JAV19455.1 GFDL01015587 JAV19458.1 GFDL01015588 JAV19457.1 CH477217 EAT47490.1 GL452008 EFN78070.1 CP012525 ALC44752.1 CCAG010002313 CCAG010002314 OUUW01000002 SPP76591.1 CH963876 EDW76775.2 GDAI01002763 JAI14840.1 NOWV01000013 RDD45829.1 AEMK02000088 DS232876 EDS26304.1 CVRI01000042 CRK95747.1 LWLT01000001 NNAY01002389 OXU21381.1 PCG80631.1 GEZM01071381 GEZM01071374 JAV65852.1 KQ979955 KYN18558.1 JH881123 ELR56450.1 GEZM01071378 GEZM01071370 JAV65859.1 AMGL01011780 AMGL01011781 AMGL01011782 BC146093 NNAY01005308 OXU16770.1 GAPW01001661 JAC11937.1 GFDF01002423 JAV11661.1 NEDP02000657 OWF55488.1 KQ762196 OAD56135.1 CH479227 EDW35306.1 CH916366 EDV97977.1 AAEX03013595 AKHW03001210 KYO43349.1 BT128180 AEE63141.1 EFN78071.1 GEHC01000248 JAV47397.1 GL442175 EFN63832.1 GABZ01006540 JAA46985.1 KJ710422 AIB53032.1 KK853123 KDR11063.1 GL888120 EGI66977.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000037510 UP000002358 UP000091820 UP000007819 UP000245320 UP000053825 UP000092445 UP000092460 UP000261681 UP000008820 UP000265300 UP000008237 UP000092553 UP000092444 UP000268350 UP000007798 UP000253843 UP000008227 UP000002320 UP000183832 UP000291000 UP000215335 UP000252040 UP000078492 UP000002356 UP000009136 UP000007646 UP000242188 UP000008744 UP000001070 UP000002254 UP000050525 UP000286640 UP000000311 UP000192223 UP000027135 UP000007755
UP000037510 UP000002358 UP000091820 UP000007819 UP000245320 UP000053825 UP000092445 UP000092460 UP000261681 UP000008820 UP000265300 UP000008237 UP000092553 UP000092444 UP000268350 UP000007798 UP000253843 UP000008227 UP000002320 UP000183832 UP000291000 UP000215335 UP000252040 UP000078492 UP000002356 UP000009136 UP000007646 UP000242188 UP000008744 UP000001070 UP000002254 UP000050525 UP000286640 UP000000311 UP000192223 UP000027135 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9JIY7
A0A2A4JFS4
A0A437AY25
I4DR67
A0A1E1WMB7
A0A2H1VAX3
+ More
S4NSI3 A0A212EPG3 A0A194PY67 A0A0N0PBK2 A0A1E1W159 A0A0L7LBD9 H9J9I3 A0A2A4K986 A0A194PS41 A0A2H1W2H8 A0A3S2LGG3 A0A194R7T8 H9JXV1 A0A437B8H1 A0A212F630 A0A2H1WGF6 A0A2H1W2M4 I4DJY6 A0A2A4K8E9 A0A194PY59 A0A2A4K851 A0A2A4K724 A0A2S2NL16 K7IZV4 A0A1A9WGA5 A0A0L7LP95 A0A0C9RY44 J9K7T8 A0A1W6EVT0 A0A2H8TD02 C1IC99 A0A2U4B980 A0A2S2QXE2 J9K623 D3TNZ8 A0A0L7QWI3 A0A1A9ZSA2 A0A1B0BCK4 A0A1Q3EW11 A0A1Q3EVW8 A0A1Q3EW78 A0A1Q3EVW7 K7IZV5 A0A384A0X1 K7IQ63 Q17L85 K7IQ61 A0A340WSB5 K7IQ62 E2C1V8 A0A0M4EIW0 A0A1B0GD38 A0A3B0JV23 B4MXK6 A0A0K8TLP0 A0A369SGB3 F1SPF0 B0XEX3 A0A287AFE9 A0A1J1I6C6 A0A452EWS2 A0A232ESQ6 A0A341CZH3 A0A2A4K8T7 A0A1Y1KWW5 A0A195E0B2 L8IIS0 A0A1Y1KWT0 W5PH63 A6H730 A0A3Q1M6R0 A0A232EEI8 A0A023EQR2 A0A1L8DYV9 G3STE9 A0A210R3A2 A0A310SN00 B4H982 B4J1A8 F1Q4D3 A0A151P351 J3JY56 E2C1V9 A0A3Q7UDM5 A0A1W7R8D5 E2ARU9 A0A1W4WJR1 K9IJ78 A0A060GL20 A0A067QNZ9 F4WFK0
S4NSI3 A0A212EPG3 A0A194PY67 A0A0N0PBK2 A0A1E1W159 A0A0L7LBD9 H9J9I3 A0A2A4K986 A0A194PS41 A0A2H1W2H8 A0A3S2LGG3 A0A194R7T8 H9JXV1 A0A437B8H1 A0A212F630 A0A2H1WGF6 A0A2H1W2M4 I4DJY6 A0A2A4K8E9 A0A194PY59 A0A2A4K851 A0A2A4K724 A0A2S2NL16 K7IZV4 A0A1A9WGA5 A0A0L7LP95 A0A0C9RY44 J9K7T8 A0A1W6EVT0 A0A2H8TD02 C1IC99 A0A2U4B980 A0A2S2QXE2 J9K623 D3TNZ8 A0A0L7QWI3 A0A1A9ZSA2 A0A1B0BCK4 A0A1Q3EW11 A0A1Q3EVW8 A0A1Q3EW78 A0A1Q3EVW7 K7IZV5 A0A384A0X1 K7IQ63 Q17L85 K7IQ61 A0A340WSB5 K7IQ62 E2C1V8 A0A0M4EIW0 A0A1B0GD38 A0A3B0JV23 B4MXK6 A0A0K8TLP0 A0A369SGB3 F1SPF0 B0XEX3 A0A287AFE9 A0A1J1I6C6 A0A452EWS2 A0A232ESQ6 A0A341CZH3 A0A2A4K8T7 A0A1Y1KWW5 A0A195E0B2 L8IIS0 A0A1Y1KWT0 W5PH63 A6H730 A0A3Q1M6R0 A0A232EEI8 A0A023EQR2 A0A1L8DYV9 G3STE9 A0A210R3A2 A0A310SN00 B4H982 B4J1A8 F1Q4D3 A0A151P351 J3JY56 E2C1V9 A0A3Q7UDM5 A0A1W7R8D5 E2ARU9 A0A1W4WJR1 K9IJ78 A0A060GL20 A0A067QNZ9 F4WFK0
PDB
1ND6
E-value=3.98705e-30,
Score=328
Ontologies
GO
GO:0003993
GO:0016021
GO:0051289
GO:0005886
GO:0051930
GO:0042131
GO:0052642
GO:0005615
GO:0042803
GO:0046085
GO:0006772
GO:0009117
GO:0012506
GO:0060168
GO:0006144
GO:0030175
GO:0008253
GO:0005549
GO:0016311
GO:0016791
GO:0006508
GO:0004181
GO:0008270
GO:0005515
GO:0005634
GO:0030145
GO:0006412
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.917123
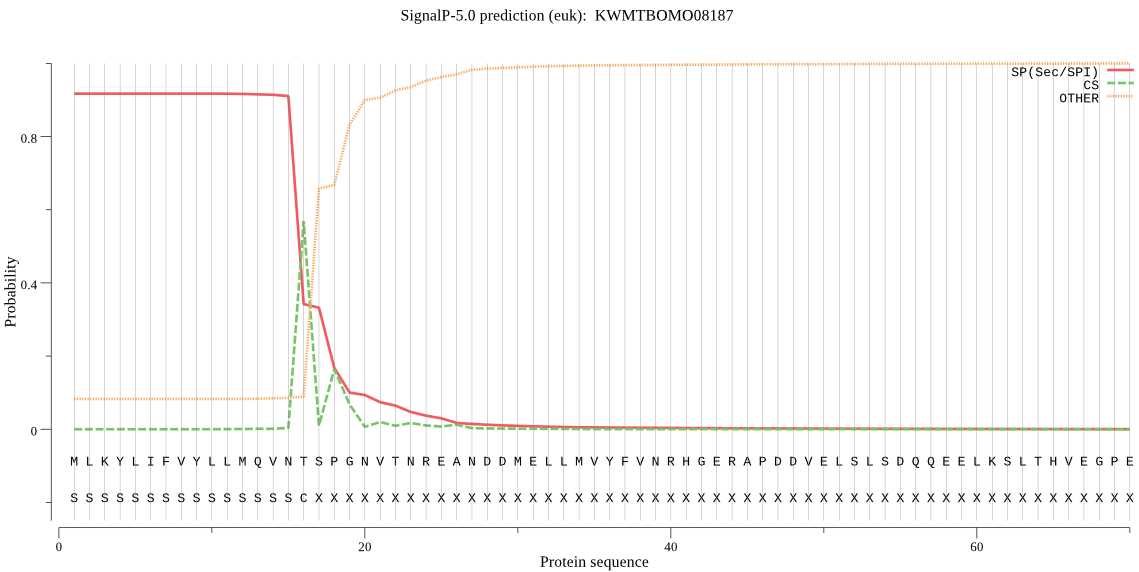
Length:
387
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01136
Exp number, first 60 AAs:
0.01055
Total prob of N-in:
0.00285
outside
1 - 387
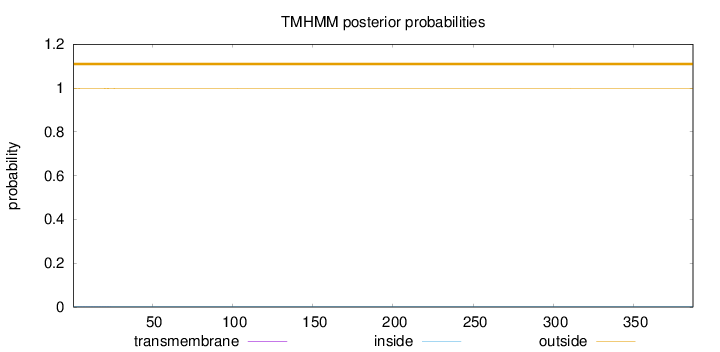
Population Genetic Test Statistics
Pi
236.671474
Theta
153.555293
Tajima's D
1.92492
CLR
0.200267
CSRT
0.868556572171391
Interpretation
Uncertain
