Gene
KWMTBOMO08183
Pre Gene Modal
BGIBMGA009486
Annotation
carboxypeptidase_B-like_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.702
Sequence
CDS
ATGAATCATTTTTTGATCCTTTCCAGATATAAATCGTACTACATATCACCTTCACACGAGAATGAGTTCCAAGCACTCGCTTCCTTGGAGAATGAGTATCAAATTGATGTTTTGACCCGGCCGATATTCGGAAGGGACGGACTGGTTCTGGTTAAGCCGGAGTACCAGGAGAGCTTCTTGGATGCTCTCGATAAATACGGCATCAAGTACAGAATTCATTTTGACGATGTGAAAGCGCAACTTGACTACGAGGACGCCATCTTGGAATTGAGAAGGAACCAAACGAGAAGTACAGAGAGATTGTCATACACCAGCTACCATGATTGGCCAGTGATTGAAGAGTACATAGACGGCATTGCTCACCGCTATCCCGATTTAGTGAAATTGGTTAATGCAGGAAGGACTTTCGAAGGGCGGAACCTCAAGTACCTGAAAATTTCCACAACAAACTTCGAAGATGCGAGCAAACCTGTTATTGTAATAGACGCAGCCATCCACGCCAGAGAATGGGCTACGATCCCTGTAGCTACTTGGGCCATCCACAAGCTAGTTGAAGACGTGACTGAGCCTGATCTGTTGAATAATTTCGATTGGATCATTTTCCCTGTTGCCAATCCCGACGGATACACGTTCTCTTTGAATACGCAACGTTTCTGGCGCAAGACCAGATCAATCGTTTCCCCCAACACAGAAAGGTGTCCGGGAGTAGACGGCAATCGAAACTATGATTTCCATTGGAACACTGTTGGTACGAGTACTAACCCTTGCGCTGAGAACTACGGCGGACCAAAGGCCTTCTCCGAGATTGAAACTAGAATAGTCCGTGATATCTTGGATGAGCACTTGTCCCGAATCAGTCTATACCTTACTATTCACAGCCATGGCAGCATGATCTTGTACCCATGGGGACACGATGGCTCTTTATCCCACAACGCTCTTGGTCTTCACACCGTTGGTATTGAAATGGCAAATGCAATCCACACTAAGGCTTTAAGCTACTTCCCAAGATATGTCGTTGGAAACGCAGCTCTCGTTTTGAGGTACGCAGCATCCGGAGCTTCTGAAGACTACGCCCATTCAGTTGGGGTGCCGCTGTCATATACCTACGAGCTTCCAGGAGGCGGTAGAGGATTCTTCTTGGATCCAATTTATATTAAACAAGTGGCTACTGAAACTTGGGAAGGCATTGTAGCCGGTGCCAGAAAATCCGAAATTTTATTTAGAGCTTAA
Protein
MNHFLILSRYKSYYISPSHENEFQALASLENEYQIDVLTRPIFGRDGLVLVKPEYQESFLDALDKYGIKYRIHFDDVKAQLDYEDAILELRRNQTRSTERLSYTSYHDWPVIEEYIDGIAHRYPDLVKLVNAGRTFEGRNLKYLKISTTNFEDASKPVIVIDAAIHAREWATIPVATWAIHKLVEDVTEPDLLNNFDWIIFPVANPDGYTFSLNTQRFWRKTRSIVSPNTERCPGVDGNRNYDFHWNTVGTSTNPCAENYGGPKAFSEIETRIVRDILDEHLSRISLYLTIHSHGSMILYPWGHDGSLSHNALGLHTVGIEMANAIHTKALSYFPRYVVGNAALVLRYAASGASEDYAHSVGVPLSYTYELPGGGRGFFLDPIYIKQVATETWEGIVAGARKSEILFRA
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9JIY5
H9JIY6
U3LUX2
C9W8K4
A0A2A4J4T7
A0A2A4J5Z1
+ More
A0A2H1VYG5 Q3T905 B1NLE5 A0A212FCH8 A0A0L7KXF0 A0A437AY42 A0A2H1VYI4 Q699Y9 Q699Y8 A0A1E1WQE4 A0A2H1VQN5 A0A0N0PBU6 A0A212FCG9 A0A194PY62 A0A1E1WL84 A0A194PM74 D2Y4U3 A0A2H1VNH7 A0A437AU59 H9J5P0 O97389 A0A194PZ08 A0A194RQH5 O97434 H9JIX7 Q9TYH8 A0A0N0PC47 A0A0N1PGB0 A0A0N1I4Q3 H9JIX6 A0A2A4JD50 Q699Z2 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 Q6H962 A0A1E1WRL9 A0A2H1W3T9 A0A2H1WCA6 Q699Z0 A0A0N1INA8 A0A2H1V2Z0 A0A3S2P6N8 A0A194PTA4 A0A212FCH2 Q6H961 Q699Z1 A0A194Q4D5 A0A212FM32 A0A0L7KWF7 A0A212F2Y7 A0A2H1WC94 A0A194PY54 B5TGP9 A0A0L7L791 A0A2H1VLT9 A0A1W4XSM1 A0A1Y1KCP4 A0A0K8V7W1 A0A1L8EHX9 A0A1A9VKY8 D3TN46 A0A0L0CSA8 A0A1I8QAI6 A0A1I8M460 A0A1B0C648 A0A1B0A6C3 A0A1A9XE82 U5EVJ7 B4HV67 A0A1L8EG86 A0A0A1XSB6 B4QKE7 Q961J8 A0A1W4UCC4 B4J297 B4KYQ6 A0A1W4UBT2 A0A0M4EJS7 B3NFD2 B4PK76 B3M5S0 A0A034V2K9 Q5QBL2 B4LDX6 B4J298 A0A0A1WK01 A0A2M3ZEP9 B4ML78 B4ML77 A0A1I8NVL3 Q9VS67 A0A1W4UNU8 A0A0A1WVY4
A0A2H1VYG5 Q3T905 B1NLE5 A0A212FCH8 A0A0L7KXF0 A0A437AY42 A0A2H1VYI4 Q699Y9 Q699Y8 A0A1E1WQE4 A0A2H1VQN5 A0A0N0PBU6 A0A212FCG9 A0A194PY62 A0A1E1WL84 A0A194PM74 D2Y4U3 A0A2H1VNH7 A0A437AU59 H9J5P0 O97389 A0A194PZ08 A0A194RQH5 O97434 H9JIX7 Q9TYH8 A0A0N0PC47 A0A0N1PGB0 A0A0N1I4Q3 H9JIX6 A0A2A4JD50 Q699Z2 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 Q6H962 A0A1E1WRL9 A0A2H1W3T9 A0A2H1WCA6 Q699Z0 A0A0N1INA8 A0A2H1V2Z0 A0A3S2P6N8 A0A194PTA4 A0A212FCH2 Q6H961 Q699Z1 A0A194Q4D5 A0A212FM32 A0A0L7KWF7 A0A212F2Y7 A0A2H1WC94 A0A194PY54 B5TGP9 A0A0L7L791 A0A2H1VLT9 A0A1W4XSM1 A0A1Y1KCP4 A0A0K8V7W1 A0A1L8EHX9 A0A1A9VKY8 D3TN46 A0A0L0CSA8 A0A1I8QAI6 A0A1I8M460 A0A1B0C648 A0A1B0A6C3 A0A1A9XE82 U5EVJ7 B4HV67 A0A1L8EG86 A0A0A1XSB6 B4QKE7 Q961J8 A0A1W4UCC4 B4J297 B4KYQ6 A0A1W4UBT2 A0A0M4EJS7 B3NFD2 B4PK76 B3M5S0 A0A034V2K9 Q5QBL2 B4LDX6 B4J298 A0A0A1WK01 A0A2M3ZEP9 B4ML78 B4ML77 A0A1I8NVL3 Q9VS67 A0A1W4UNU8 A0A0A1WVY4
Pubmed
EMBL
BABH01022994
BABH01022995
JX865781
AGL42356.1
FJ210817
ACN69214.1
+ More
NWSH01002977 PCG67157.1 PCG67156.1 ODYU01005225 SOQ45875.1 AM085495 CAJ30028.1 EF600060 ABU98625.1 AGBW02009192 OWR51430.1 JTDY01004612 KOB67948.1 RSAL01000278 RVE43076.1 SOQ45873.1 AY512677 AAS82586.1 AY512678 AAS82587.1 GDQN01002013 JAT89041.1 ODYU01003862 SOQ43155.1 KQ460778 KPJ12424.1 OWR51431.1 KQ459585 KPI98272.1 GDQN01003319 JAT87735.1 KQ459604 KPI92215.1 GU323795 ADA83700.1 ODYU01003485 SOQ42347.1 RSAL01000517 RVE41479.1 BABH01024959 BABH01024960 AJ005177 CAA06418.1 KPI98273.1 KQ460045 KPJ18276.1 AJ005178 CAA06419.1 BABH01022991 AJ005176 CAA06417.1 KPJ12422.1 KPJ12423.1 KQ461189 KPJ07295.1 BABH01022992 NWSH01002042 PCG69323.1 AY512674 AAS82583.1 GDQN01005910 JAT85144.1 KPJ12425.1 GU325786 ADB96010.1 AJ626862 CAF25189.2 GDQN01001497 JAT89557.1 ODYU01006127 SOQ47708.1 ODYU01007689 SOQ50698.1 AY512676 AAS82585.1 KPJ07296.1 ODYU01000433 SOQ35191.1 RVE43075.1 KQ459595 KPI95994.1 OWR51432.1 AJ626863 CAF25190.1 AY512675 AAS82584.1 KPI98270.1 AGBW02007655 OWR54814.1 JTDY01005092 KOB67379.1 AGBW02010665 OWR48083.1 SOQ50701.1 KPI98271.1 EU924507 ACH82086.1 JTDY01002459 KOB71372.1 ODYU01003089 SOQ41402.1 GEZM01092428 JAV56557.1 GDHF01017649 JAI34665.1 GFDG01000545 JAV18254.1 CCAG010004976 EZ422848 ADD19124.1 JRES01000080 KNC34299.1 JXJN01026429 GANO01000996 JAB58875.1 CH480817 EDW50838.1 GFDG01001145 JAV17654.1 GBXI01000083 JAD14209.1 CM000363 CM002912 EDX09570.1 KMY98094.1 AE014296 AY051552 AAF50560.2 AAK92976.1 CH916366 EDV97048.1 CH933809 EDW17770.1 CP012525 ALC43322.1 CH954178 EDV50474.1 CM000159 EDW93756.1 CH902618 EDV40704.1 GAKP01022233 JAC36719.1 AY752799 AAV84212.1 CH940647 EDW68999.1 EDV97049.1 GBXI01014928 JAC99363.1 GGFM01006222 MBW26973.1 CH963847 EDW73136.1 EDW73135.1 AY071045 AAF50561.2 AAL48667.1 GBXI01011699 JAD02593.1
NWSH01002977 PCG67157.1 PCG67156.1 ODYU01005225 SOQ45875.1 AM085495 CAJ30028.1 EF600060 ABU98625.1 AGBW02009192 OWR51430.1 JTDY01004612 KOB67948.1 RSAL01000278 RVE43076.1 SOQ45873.1 AY512677 AAS82586.1 AY512678 AAS82587.1 GDQN01002013 JAT89041.1 ODYU01003862 SOQ43155.1 KQ460778 KPJ12424.1 OWR51431.1 KQ459585 KPI98272.1 GDQN01003319 JAT87735.1 KQ459604 KPI92215.1 GU323795 ADA83700.1 ODYU01003485 SOQ42347.1 RSAL01000517 RVE41479.1 BABH01024959 BABH01024960 AJ005177 CAA06418.1 KPI98273.1 KQ460045 KPJ18276.1 AJ005178 CAA06419.1 BABH01022991 AJ005176 CAA06417.1 KPJ12422.1 KPJ12423.1 KQ461189 KPJ07295.1 BABH01022992 NWSH01002042 PCG69323.1 AY512674 AAS82583.1 GDQN01005910 JAT85144.1 KPJ12425.1 GU325786 ADB96010.1 AJ626862 CAF25189.2 GDQN01001497 JAT89557.1 ODYU01006127 SOQ47708.1 ODYU01007689 SOQ50698.1 AY512676 AAS82585.1 KPJ07296.1 ODYU01000433 SOQ35191.1 RVE43075.1 KQ459595 KPI95994.1 OWR51432.1 AJ626863 CAF25190.1 AY512675 AAS82584.1 KPI98270.1 AGBW02007655 OWR54814.1 JTDY01005092 KOB67379.1 AGBW02010665 OWR48083.1 SOQ50701.1 KPI98271.1 EU924507 ACH82086.1 JTDY01002459 KOB71372.1 ODYU01003089 SOQ41402.1 GEZM01092428 JAV56557.1 GDHF01017649 JAI34665.1 GFDG01000545 JAV18254.1 CCAG010004976 EZ422848 ADD19124.1 JRES01000080 KNC34299.1 JXJN01026429 GANO01000996 JAB58875.1 CH480817 EDW50838.1 GFDG01001145 JAV17654.1 GBXI01000083 JAD14209.1 CM000363 CM002912 EDX09570.1 KMY98094.1 AE014296 AY051552 AAF50560.2 AAK92976.1 CH916366 EDV97048.1 CH933809 EDW17770.1 CP012525 ALC43322.1 CH954178 EDV50474.1 CM000159 EDW93756.1 CH902618 EDV40704.1 GAKP01022233 JAC36719.1 AY752799 AAV84212.1 CH940647 EDW68999.1 EDV97049.1 GBXI01014928 JAC99363.1 GGFM01006222 MBW26973.1 CH963847 EDW73136.1 EDW73135.1 AY071045 AAF50561.2 AAL48667.1 GBXI01011699 JAD02593.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000283053
UP000053240
+ More
UP000053268 UP000192223 UP000078200 UP000092444 UP000037069 UP000095300 UP000095301 UP000092460 UP000092445 UP000092443 UP000001292 UP000000304 UP000000803 UP000192221 UP000001070 UP000009192 UP000092553 UP000008711 UP000002282 UP000007801 UP000008792 UP000007798
UP000053268 UP000192223 UP000078200 UP000092444 UP000037069 UP000095300 UP000095301 UP000092460 UP000092445 UP000092443 UP000001292 UP000000304 UP000000803 UP000192221 UP000001070 UP000009192 UP000092553 UP000008711 UP000002282 UP000007801 UP000008792 UP000007798
Interpro
Gene 3D
ProteinModelPortal
H9JIY5
H9JIY6
U3LUX2
C9W8K4
A0A2A4J4T7
A0A2A4J5Z1
+ More
A0A2H1VYG5 Q3T905 B1NLE5 A0A212FCH8 A0A0L7KXF0 A0A437AY42 A0A2H1VYI4 Q699Y9 Q699Y8 A0A1E1WQE4 A0A2H1VQN5 A0A0N0PBU6 A0A212FCG9 A0A194PY62 A0A1E1WL84 A0A194PM74 D2Y4U3 A0A2H1VNH7 A0A437AU59 H9J5P0 O97389 A0A194PZ08 A0A194RQH5 O97434 H9JIX7 Q9TYH8 A0A0N0PC47 A0A0N1PGB0 A0A0N1I4Q3 H9JIX6 A0A2A4JD50 Q699Z2 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 Q6H962 A0A1E1WRL9 A0A2H1W3T9 A0A2H1WCA6 Q699Z0 A0A0N1INA8 A0A2H1V2Z0 A0A3S2P6N8 A0A194PTA4 A0A212FCH2 Q6H961 Q699Z1 A0A194Q4D5 A0A212FM32 A0A0L7KWF7 A0A212F2Y7 A0A2H1WC94 A0A194PY54 B5TGP9 A0A0L7L791 A0A2H1VLT9 A0A1W4XSM1 A0A1Y1KCP4 A0A0K8V7W1 A0A1L8EHX9 A0A1A9VKY8 D3TN46 A0A0L0CSA8 A0A1I8QAI6 A0A1I8M460 A0A1B0C648 A0A1B0A6C3 A0A1A9XE82 U5EVJ7 B4HV67 A0A1L8EG86 A0A0A1XSB6 B4QKE7 Q961J8 A0A1W4UCC4 B4J297 B4KYQ6 A0A1W4UBT2 A0A0M4EJS7 B3NFD2 B4PK76 B3M5S0 A0A034V2K9 Q5QBL2 B4LDX6 B4J298 A0A0A1WK01 A0A2M3ZEP9 B4ML78 B4ML77 A0A1I8NVL3 Q9VS67 A0A1W4UNU8 A0A0A1WVY4
A0A2H1VYG5 Q3T905 B1NLE5 A0A212FCH8 A0A0L7KXF0 A0A437AY42 A0A2H1VYI4 Q699Y9 Q699Y8 A0A1E1WQE4 A0A2H1VQN5 A0A0N0PBU6 A0A212FCG9 A0A194PY62 A0A1E1WL84 A0A194PM74 D2Y4U3 A0A2H1VNH7 A0A437AU59 H9J5P0 O97389 A0A194PZ08 A0A194RQH5 O97434 H9JIX7 Q9TYH8 A0A0N0PC47 A0A0N1PGB0 A0A0N1I4Q3 H9JIX6 A0A2A4JD50 Q699Z2 A0A1E1WDT2 A0A0N1IDD0 D3JXJ8 Q6H962 A0A1E1WRL9 A0A2H1W3T9 A0A2H1WCA6 Q699Z0 A0A0N1INA8 A0A2H1V2Z0 A0A3S2P6N8 A0A194PTA4 A0A212FCH2 Q6H961 Q699Z1 A0A194Q4D5 A0A212FM32 A0A0L7KWF7 A0A212F2Y7 A0A2H1WC94 A0A194PY54 B5TGP9 A0A0L7L791 A0A2H1VLT9 A0A1W4XSM1 A0A1Y1KCP4 A0A0K8V7W1 A0A1L8EHX9 A0A1A9VKY8 D3TN46 A0A0L0CSA8 A0A1I8QAI6 A0A1I8M460 A0A1B0C648 A0A1B0A6C3 A0A1A9XE82 U5EVJ7 B4HV67 A0A1L8EG86 A0A0A1XSB6 B4QKE7 Q961J8 A0A1W4UCC4 B4J297 B4KYQ6 A0A1W4UBT2 A0A0M4EJS7 B3NFD2 B4PK76 B3M5S0 A0A034V2K9 Q5QBL2 B4LDX6 B4J298 A0A0A1WK01 A0A2M3ZEP9 B4ML78 B4ML77 A0A1I8NVL3 Q9VS67 A0A1W4UNU8 A0A0A1WVY4
PDB
2C1C
E-value=9.64294e-118,
Score=1083
Ontologies
GO
Topology
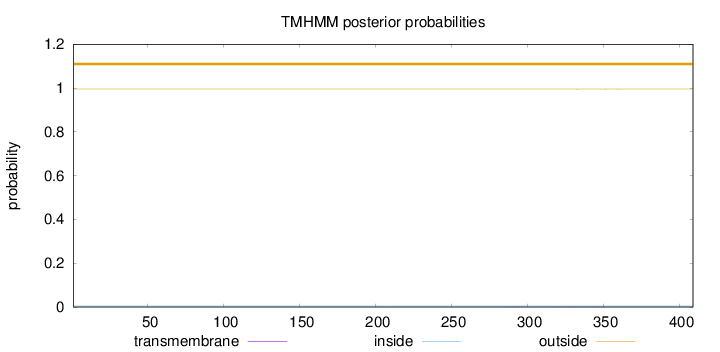
Length:
409
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02217
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00425
outside
1 - 409
Population Genetic Test Statistics
Pi
50.875966
Theta
141.111974
Tajima's D
-0.833376
CLR
298.017538
CSRT
0.169491525423729
Interpretation
Possibly Positive selection
