Gene
KWMTBOMO08171
Annotation
PREDICTED:_zinc_finger_BED_domain-containing_protein_1-like_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 2.942
Sequence
CDS
ATGGGACCTAAGAAAAAAAGTTCCGTTTGGCAGTTCTTTAACAAAATTGACGATAAGAAAACGAAATGCAAACTATGTCAAAAGGAAATTAAGTCTGCTGGAAACACTACGAACTTAATTGGACATATTCGTAATGTTCATAAGGCTGCTTATCTCGAAATTCAGCCGAAACAAACTACATTCAATGCAGTAACTACTACAGACGAAATATCGAATCCTCTGGATCTGGATACCAAAGTTACGCCTTATAATAAAATGGATGATGGAGTACTTCAGTTTCAACCTGTTCCAAGTTGCAGTGGCAGTCAAAGATCCGGTGCACCGTTACCAAAAACCGTAAATGAAACCATTGATTCAGACTTAGACGAAAACGTTACACCTTTAAAACGCCAAAGAACAATACAGGCCGCTTTTAAAGATATTTCCTCTTACTCAGAATCTGGAGCGAAGACAAGGAAACTGAACAACTGTTTGTTATTCATGATTTGCAAAGACCATCAGCCATTCTCAATCGTTGAAAACGAAGGCTTCAAAACTCTAATGAAAACAGTTGCTCCTCAATATAAGCTCCCGAGTAGAACAACTCTAAGACGCTGGCTGGACGATAAATATGAAGTAATTTCAGAAACAATTAAGAAAATGTTGTCCTCTATAGAAGATATAACCCTAACAACAGATATTTGGTCGGATACCTTGAATATGAAAAGTTTTTTGGGCATCACAGCACATTTCGGAATAGACATTGAACTATATTCAGTCACCTTAGGTGTGTACGAATTGGATGAAAGACATACATCAGATTATCTTTCTGAAATTCTCTTAAAGGTATGTCAAGACTGGAAAATCGATCAAGAGAATGTAACAGCTGTTGTTACTGACAACGCAGCCAATATAGTTAAGGCGATCGAATTAGCGTTTGGTAGAAGGAAACACATCCCATGTTTTGCCCATACTCTGAATTTGGTGGCTGAAGGCACAATGATATGTATTGAGTGGAGAAACATTGTTTCTAAAGTAAAATCAATAGTGACTTGGTTTAAACAGAGTTGCATTGCTAGTGATGAATTACGTAAAGCTACTGCTGCGGCTACGGAGTCTTCTGCTGGAGCAGCTGAGGTAAAATTAATACAAAGCGTGGATACTCGCTGGAACAGTACATATTATATGTTACAAAGATTTCTAGAACTCCGTAGTGTCATAAATGATATTCTCTTCCGCCACCCTAGAGCACCTGCAATGCTGAGTGCATCGGAAATATCTACTGTATCGTCTGTCATCATGGTATTGCGACCCTTAGAAGCGGCTACAAAAGAAATTTCAGGTGATAAATACTGCACAAGTAGTAAAATTATTCCCTTAGTTCGTTGCATGCTCTCTAAAATAACTTCAGCTGTTATAGAAGATCCCGTAGCGAAAGAGGTGCAGAAACTTGCTATTAATGAAATAAATAAAAGAATGGGTTCAATAGAACATGTGAATGCTCTGGCTATCGCAAGTATTTTAGATCCACGGTTTAAAAGAATGCATTTCAATGATCCGATAGCTTGTTCGAATGCTGTTACGAAAATTAAAGATTTAATGAAGAAGAATTTGCATAGCAATGAAGAACCTGAGTCCGATTCTGACAAGTCCGATAAAAATGAAGAGGCCTTTTCATTATGGACTGATCACCATAAGTTAGTGCATCGTAACTGGAAAATTAACAAATCTGAAGACGTTTTGTCTGACGAACTTTCCGTTTATCTCCGAACTCCTGTTGGAAGACTTAAAGAAAATCCGTTAGAAATTTGGTGA
Protein
MGPKKKSSVWQFFNKIDDKKTKCKLCQKEIKSAGNTTNLIGHIRNVHKAAYLEIQPKQTTFNAVTTTDEISNPLDLDTKVTPYNKMDDGVLQFQPVPSCSGSQRSGAPLPKTVNETIDSDLDENVTPLKRQRTIQAAFKDISSYSESGAKTRKLNNCLLFMICKDHQPFSIVENEGFKTLMKTVAPQYKLPSRTTLRRWLDDKYEVISETIKKMLSSIEDITLTTDIWSDTLNMKSFLGITAHFGIDIELYSVTLGVYELDERHTSDYLSEILLKVCQDWKIDQENVTAVVTDNAANIVKAIELAFGRRKHIPCFAHTLNLVAEGTMICIEWRNIVSKVKSIVTWFKQSCIASDELRKATAAATESSAGAAEVKLIQSVDTRWNSTYYMLQRFLELRSVINDILFRHPRAPAMLSASEISTVSSVIMVLRPLEAATKEISGDKYCTSSKIIPLVRCMLSKITSAVIEDPVAKEVQKLAINEINKRMGSIEHVNALAIASILDPRFKRMHFNDPIACSNAVTKIKDLMKKNLHSNEEPESDSDKSDKNEEAFSLWTDHHKLVHRNWKINKSEDVLSDELSVYLRTPVGRLKENPLEIW
Summary
Uniprot
J9LDY8
A0A1W4WLZ1
J9L2Y9
J9M7K2
A0A1E1WN08
J9KF88
+ More
J9M2M5 A0A151IJ50 E2AN53 J9M5N4 J9KNP5 A0A151J3A2 A0A151ICL6 A0A0J7K4Y0 J9LFC3 E2C2R7 A0A195DX32 A0A151K1W2 A0A151J8J8 A0A026X0S0 X1WXE1 A0A151IVN1 A0A3L8DVM9 A0A2S2QDH1 A0A026WKR9 A0A2S2R3S9 J9LSA5 A0A0J7KSL2 A0A151IK53 A0A026VWT7 A0A437AZ90 A0A224XKI7 Q8T648 J9LEV9 A0A158P559 A0A3P8RVY2 X1X7U1 T1JXM7 J9LVM0 A0A3B1JDP8 A0A2S2QCJ4 E2A4Y2 A0A232EFR6 J9LS52 A0A1X7UCW0 A0A3B3BJP2 J9KBT0 X1WTG7 G3PR68 J9KJS4 A0A1X7TGY6 A0A1W4XFU6 X1XCD4 A0A3Q3GI05 T1K5U2 A0A1X7UJA9 A0A0A1XSV1 A0A2S2PB02 A0A0K2UYR9 A0A1X7UFA9 A0A2A4JMY2 J9KDG5 A0A2A4JLR2 A0A151IU19 X1XTH2 A0A034W0I4
J9M2M5 A0A151IJ50 E2AN53 J9M5N4 J9KNP5 A0A151J3A2 A0A151ICL6 A0A0J7K4Y0 J9LFC3 E2C2R7 A0A195DX32 A0A151K1W2 A0A151J8J8 A0A026X0S0 X1WXE1 A0A151IVN1 A0A3L8DVM9 A0A2S2QDH1 A0A026WKR9 A0A2S2R3S9 J9LSA5 A0A0J7KSL2 A0A151IK53 A0A026VWT7 A0A437AZ90 A0A224XKI7 Q8T648 J9LEV9 A0A158P559 A0A3P8RVY2 X1X7U1 T1JXM7 J9LVM0 A0A3B1JDP8 A0A2S2QCJ4 E2A4Y2 A0A232EFR6 J9LS52 A0A1X7UCW0 A0A3B3BJP2 J9KBT0 X1WTG7 G3PR68 J9KJS4 A0A1X7TGY6 A0A1W4XFU6 X1XCD4 A0A3Q3GI05 T1K5U2 A0A1X7UJA9 A0A0A1XSV1 A0A2S2PB02 A0A0K2UYR9 A0A1X7UFA9 A0A2A4JMY2 J9KDG5 A0A2A4JLR2 A0A151IU19 X1XTH2 A0A034W0I4
EMBL
ABLF02010709
ABLF02009488
ABLF02008875
GDQN01002674
JAT88380.1
ABLF02030582
+ More
ABLF02006015 KQ977395 KYN02984.1 GL441080 EFN65135.1 ABLF02001734 ABLF02023260 KQ980290 KYN16919.1 KQ978037 KYM97822.1 LBMM01014067 KMQ85349.1 ABLF02011645 ABLF02044591 ABLF02050984 ABLF02055777 GL452203 EFN77752.1 KQ980204 KYN17277.1 LKEX01009590 KYN50115.1 KQ979507 KYN21183.1 KK107050 EZA61658.1 ABLF02026056 ABLF02057203 KQ980913 KYN11465.1 QOIP01000003 RLU24471.1 GGMS01006357 MBY75560.1 KK107161 EZA56605.1 GGMS01015207 MBY84410.1 ABLF02017802 LBMM01003701 KMQ93224.1 KQ977255 KYN04655.1 KK107725 EZA47986.1 RSAL01000257 RVE43413.1 GFTR01007466 JAW08960.1 AF486809 AAL93203.1 ABLF02018406 CAEY01000199 ABLF02012532 CAEY01000832 ABLF02006624 ABLF02006626 GGMS01006230 MBY75433.1 GL436768 EFN71507.1 NNAY01004948 OXU17200.1 ABLF02021118 ABLF02014501 ABLF02011646 ABLF02013863 ABLF02067360 CAEY01001590 GBXI01000181 JAD14111.1 GGMR01013943 MBY26562.1 HACA01026053 CDW43414.1 NWSH01001073 PCG72760.1 ABLF02055338 PCG72759.1 KQ980971 KYN10910.1 ABLF02011642 GAKP01011699 JAC47253.1
ABLF02006015 KQ977395 KYN02984.1 GL441080 EFN65135.1 ABLF02001734 ABLF02023260 KQ980290 KYN16919.1 KQ978037 KYM97822.1 LBMM01014067 KMQ85349.1 ABLF02011645 ABLF02044591 ABLF02050984 ABLF02055777 GL452203 EFN77752.1 KQ980204 KYN17277.1 LKEX01009590 KYN50115.1 KQ979507 KYN21183.1 KK107050 EZA61658.1 ABLF02026056 ABLF02057203 KQ980913 KYN11465.1 QOIP01000003 RLU24471.1 GGMS01006357 MBY75560.1 KK107161 EZA56605.1 GGMS01015207 MBY84410.1 ABLF02017802 LBMM01003701 KMQ93224.1 KQ977255 KYN04655.1 KK107725 EZA47986.1 RSAL01000257 RVE43413.1 GFTR01007466 JAW08960.1 AF486809 AAL93203.1 ABLF02018406 CAEY01000199 ABLF02012532 CAEY01000832 ABLF02006624 ABLF02006626 GGMS01006230 MBY75433.1 GL436768 EFN71507.1 NNAY01004948 OXU17200.1 ABLF02021118 ABLF02014501 ABLF02011646 ABLF02013863 ABLF02067360 CAEY01001590 GBXI01000181 JAD14111.1 GGMR01013943 MBY26562.1 HACA01026053 CDW43414.1 NWSH01001073 PCG72760.1 ABLF02055338 PCG72759.1 KQ980971 KYN10910.1 ABLF02011642 GAKP01011699 JAC47253.1
Proteomes
Interpro
ProteinModelPortal
J9LDY8
A0A1W4WLZ1
J9L2Y9
J9M7K2
A0A1E1WN08
J9KF88
+ More
J9M2M5 A0A151IJ50 E2AN53 J9M5N4 J9KNP5 A0A151J3A2 A0A151ICL6 A0A0J7K4Y0 J9LFC3 E2C2R7 A0A195DX32 A0A151K1W2 A0A151J8J8 A0A026X0S0 X1WXE1 A0A151IVN1 A0A3L8DVM9 A0A2S2QDH1 A0A026WKR9 A0A2S2R3S9 J9LSA5 A0A0J7KSL2 A0A151IK53 A0A026VWT7 A0A437AZ90 A0A224XKI7 Q8T648 J9LEV9 A0A158P559 A0A3P8RVY2 X1X7U1 T1JXM7 J9LVM0 A0A3B1JDP8 A0A2S2QCJ4 E2A4Y2 A0A232EFR6 J9LS52 A0A1X7UCW0 A0A3B3BJP2 J9KBT0 X1WTG7 G3PR68 J9KJS4 A0A1X7TGY6 A0A1W4XFU6 X1XCD4 A0A3Q3GI05 T1K5U2 A0A1X7UJA9 A0A0A1XSV1 A0A2S2PB02 A0A0K2UYR9 A0A1X7UFA9 A0A2A4JMY2 J9KDG5 A0A2A4JLR2 A0A151IU19 X1XTH2 A0A034W0I4
J9M2M5 A0A151IJ50 E2AN53 J9M5N4 J9KNP5 A0A151J3A2 A0A151ICL6 A0A0J7K4Y0 J9LFC3 E2C2R7 A0A195DX32 A0A151K1W2 A0A151J8J8 A0A026X0S0 X1WXE1 A0A151IVN1 A0A3L8DVM9 A0A2S2QDH1 A0A026WKR9 A0A2S2R3S9 J9LSA5 A0A0J7KSL2 A0A151IK53 A0A026VWT7 A0A437AZ90 A0A224XKI7 Q8T648 J9LEV9 A0A158P559 A0A3P8RVY2 X1X7U1 T1JXM7 J9LVM0 A0A3B1JDP8 A0A2S2QCJ4 E2A4Y2 A0A232EFR6 J9LS52 A0A1X7UCW0 A0A3B3BJP2 J9KBT0 X1WTG7 G3PR68 J9KJS4 A0A1X7TGY6 A0A1W4XFU6 X1XCD4 A0A3Q3GI05 T1K5U2 A0A1X7UJA9 A0A0A1XSV1 A0A2S2PB02 A0A0K2UYR9 A0A1X7UFA9 A0A2A4JMY2 J9KDG5 A0A2A4JLR2 A0A151IU19 X1XTH2 A0A034W0I4
PDB
6DX0
E-value=4.18294e-14,
Score=191
Ontologies
GO
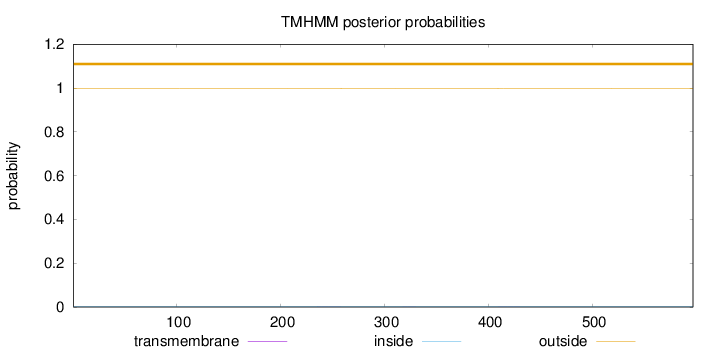
Topology
Length:
597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02103
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00168
outside
1 - 597
Population Genetic Test Statistics
Pi
215.500937
Theta
212.011977
Tajima's D
0.040815
CLR
0.000861
CSRT
0.383380830958452
Interpretation
Uncertain
