Gene
KWMTBOMO08168
Pre Gene Modal
BGIBMGA014381
Annotation
PREDICTED:_apoptosis-inducing_factor_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.919
Sequence
CDS
ATGGAACACGATAAGAAAGCAAAGGTGTTCTTCATAAGCAAGGAAGACTGTCTACCGTACATTAGACTTCCAATGTCGAAGCAAATGTGGTGGAACCCCGATCCGCCTGACATCAAATCCCTCAACTATTTAGAAGACGGAAAGAGACGCACCATGTACCTAGCGGATTGTTCGAAGTTCATGGATCCGGTGAAGTTTTACCGGACTAAGACCGGACCGGCCGTGTCCGTGGCGACCGGATGGTGCGTGCTGAGGGTGGACGCAGACGAGCACGTTGTCTACGTGAAGACAATGTGCGGCGAGCAGCCTATATACTACGAGAGGTGCTTACTGGCTCCTGGTTCCAAACCGAAGAGTTTGAGCGTGTTCAAGTCGGCGCCCAAAGCGGTCCGGCAGAAGGTGTGCGCGCTGCGGACCGTGCGCGACCTGGAGCTGGCGCAGCGCGGGGTGCGGCGCGCGCGACACGTGGTCGTGCTGGGCGGCGGCTGTCTGGGCACCGAGCTGGCCTGGCATCTGGGGAGGATGAATCAGCTCATCGAACGTAAGCCCGATGAGGAGCCGCTGCAGATAGTCCACGTCTTCAAAGACAAAGGCATCATGGCTGGAGTCATACCAGACTATCTCGGCGAGTGGGCCACTGAGAAGATCAAATGCGAAGGCGTCACCGTAATGCCTAAAACGCAAGTGTACGACGCGTTCGAGACGCCCGACGGGAGGGTCCAGTTGACGCTGTCTAACGGAACTTCCCTGGTCACGGACTACGTGCTAGTGGCGGTGGGAGCCGAACCGCGAATGGAACTGGCCGAACCCTCGTTTCTGGAGCAGGACCCGGTCAACGGCGGATTCCTGGTCAACACAGAACTGGAAGCTAGAACACATCTATACGTTGCCGGAGACGCGGCCAGTGTGTACTCGCAGTGGCGCGACGTCCGCTTCCGGGACGAGCACTACGACACGGCCGAGCGGCTCGGTCTCATCGCCGGCGCCAACATGACGGGCCACTGGCTCGCCTGCAACCTCGAGCCGCACTACTGGCTCCGGCTCGGAGACGCGCTGCAGATGGAGGTGGTGGGTGAAGTGGGGGCTTGTCTCCCGACGATCGCGTTGTTCAAGCAATGCGCTCCTGAAGAGGTCCCCAAAAAGACGGAGACCCCCGCGGGTGATGGCGAAAAACCTTGCTACAAAAAGTCGGAGGAGTACCAGAACCGATACAAGCGAGGCATCGTGTTCTACCTGCGCGACGAGACCGTGGTGGGGCTGGTCTTCTGGAACATGCCGCCCATCGACGACAGGAGGGACGTGGCCACCGAGATCCTTCGCGCTCGACCCTCGTACAAAGATATAAACCTGCTAGCTGAACTCCTGGGCTTCGCAGAGACGCAGTGCGTTTACAAAACGCACGAGCAACTCAAGGAACCTGGTCCTTGCATTCGAAACTGGAGAGAATTTTGA
Protein
MEHDKKAKVFFISKEDCLPYIRLPMSKQMWWNPDPPDIKSLNYLEDGKRRTMYLADCSKFMDPVKFYRTKTGPAVSVATGWCVLRVDADEHVVYVKTMCGEQPIYYERCLLAPGSKPKSLSVFKSAPKAVRQKVCALRTVRDLELAQRGVRRARHVVVLGGGCLGTELAWHLGRMNQLIERKPDEEPLQIVHVFKDKGIMAGVIPDYLGEWATEKIKCEGVTVMPKTQVYDAFETPDGRVQLTLSNGTSLVTDYVLVAVGAEPRMELAEPSFLEQDPVNGGFLVNTELEARTHLYVAGDAASVYSQWRDVRFRDEHYDTAERLGLIAGANMTGHWLACNLEPHYWLRLGDALQMEVVGEVGACLPTIALFKQCAPEEVPKKTETPAGDGEKPCYKKSEEYQNRYKRGIVFYLRDETVVGLVFWNMPPIDDRRDVATEILRARPSYKDINLLAELLGFAETQCVYKTHEQLKEPGPCIRNWREF
Summary
Uniprot
E9JEG6
A0A0N1ICV2
A0A212F5H5
A0A194PY77
A0A2W1BF90
A0A2H1WZL8
+ More
A0A2M3Z7Q7 A0A2M4AAJ3 W5JKX5 A0A182F616 A0A1Q3FED4 A0A2M3Z799 B0W7M7 A0A2M4BHK9 A0A2M4BHD0 A0A1W4WGV9 A0A1Q3FE57 A0A2M4AAQ2 Q17JG7 A0A182UAE0 A0A1S4GXS5 A0A182WUA0 A0A0C9QS22 A0A182L1R2 Q7Q3U0 A0A182HUW8 A0A182JR64 A0A1Y1LC16 A0A182GSK7 A0A182UYX4 A0A0K8TNI5 E0W028 A0A182RC80 A0A182Y019 A0A443RNN7 A0A087T8Q9 A0A2S2PEQ0 A0A3T0JLN2 A0A2H8TVU6 A0A2S2QWH0 A0A2P2HWW7 A0A182P4V7 A0A423S9X6 A0A087TMI7 A0A1S3LPT9 A0A443SRS7 A0A3S1B2K8 A0A084VND9 A0A1B1GE10 A0A1W4ZEJ7 A0A3B3RGD9 T1J3Z2 A0A1W4ZMW1 A0A1S3LQ11 A0A0P5M571 A0A1S3LQ57 A0A0P4Z3H5 A0A3B3RGH1 A0A1S3LQU2 A0A0P5INF4 A0A1W4ZEJ8 A0A1W4ZN17 T1KHA7 A0A3B3RG57 A0A1S3LPN4 H9GEN1 A0A0P5FRE2 A0A0P4Z9V0 A0A0K8VG98 A0A1S3LPT3 A0A1S3LQ52 A0A1J1IVD3 A0A0P4WPP2 A0A1S3LQ07 A0A1S3LQT7 A0A2C9JBH3 A0A0P5Q211 H2ZVB3 A0A0P5SLX2 A0A0P5BIH7 A0A0P4YA23 A0A182YTT5 A0A0P5HWQ5 A0A0P4WSA2 A0A0N8C166 A0A060WQQ5 A0A0P5ATI5 A0A182QQX4 A0A0P5AQW0 H2ZVB2 A0A0P5WI87 A0A0P5QDI6 A0A0P5BBX7 A0A0P5H8Z7 A0A0T6B2J4 A0A0P5DD02 A0A091KUC9 A0A0P5BJ72
A0A2M3Z7Q7 A0A2M4AAJ3 W5JKX5 A0A182F616 A0A1Q3FED4 A0A2M3Z799 B0W7M7 A0A2M4BHK9 A0A2M4BHD0 A0A1W4WGV9 A0A1Q3FE57 A0A2M4AAQ2 Q17JG7 A0A182UAE0 A0A1S4GXS5 A0A182WUA0 A0A0C9QS22 A0A182L1R2 Q7Q3U0 A0A182HUW8 A0A182JR64 A0A1Y1LC16 A0A182GSK7 A0A182UYX4 A0A0K8TNI5 E0W028 A0A182RC80 A0A182Y019 A0A443RNN7 A0A087T8Q9 A0A2S2PEQ0 A0A3T0JLN2 A0A2H8TVU6 A0A2S2QWH0 A0A2P2HWW7 A0A182P4V7 A0A423S9X6 A0A087TMI7 A0A1S3LPT9 A0A443SRS7 A0A3S1B2K8 A0A084VND9 A0A1B1GE10 A0A1W4ZEJ7 A0A3B3RGD9 T1J3Z2 A0A1W4ZMW1 A0A1S3LQ11 A0A0P5M571 A0A1S3LQ57 A0A0P4Z3H5 A0A3B3RGH1 A0A1S3LQU2 A0A0P5INF4 A0A1W4ZEJ8 A0A1W4ZN17 T1KHA7 A0A3B3RG57 A0A1S3LPN4 H9GEN1 A0A0P5FRE2 A0A0P4Z9V0 A0A0K8VG98 A0A1S3LPT3 A0A1S3LQ52 A0A1J1IVD3 A0A0P4WPP2 A0A1S3LQ07 A0A1S3LQT7 A0A2C9JBH3 A0A0P5Q211 H2ZVB3 A0A0P5SLX2 A0A0P5BIH7 A0A0P4YA23 A0A182YTT5 A0A0P5HWQ5 A0A0P4WSA2 A0A0N8C166 A0A060WQQ5 A0A0P5ATI5 A0A182QQX4 A0A0P5AQW0 H2ZVB2 A0A0P5WI87 A0A0P5QDI6 A0A0P5BBX7 A0A0P5H8Z7 A0A0T6B2J4 A0A0P5DD02 A0A091KUC9 A0A0P5BJ72
Pubmed
EMBL
BABH01040393
GQ426283
ADM32509.1
KQ460889
KPJ11050.1
AGBW02010196
+ More
OWR48986.1 KQ459585 KPI98287.1 KZ150088 PZC73742.1 ODYU01011849 SOQ57824.1 GGFM01003801 MBW24552.1 GGFK01004431 MBW37752.1 ADMH02000877 ETN64791.1 GFDL01009132 JAV25913.1 GGFM01003567 MBW24318.1 DS231854 EDS38062.1 GGFJ01003300 MBW52441.1 GGFJ01003301 MBW52442.1 GFDL01009247 JAV25798.1 GGFK01004550 MBW37871.1 CH477232 EAT46873.1 AAAB01008964 GBYB01003442 JAG73209.1 EAA12325.4 APCN01002480 GEZM01061486 JAV70438.1 JXUM01017001 KQ560472 KXJ82231.1 GDAI01001905 JAI15698.1 DS235855 EEB18984.1 NCKU01000159 RWS16828.1 KK113991 KFM61498.1 GGMR01014767 MBY27386.1 MH393923 AZV23876.1 GFXV01006234 MBW18039.1 GGMS01012908 MBY82111.1 IACF01000468 LAB66242.1 QCYY01004418 ROT61031.1 KK115903 KFM66326.1 NCKV01000613 RWS30165.1 RQTK01000929 RUS73496.1 ATLV01014737 KE524984 KFB39483.1 KX096891 ANQ68316.1 JH431832 GDIQ01169164 JAK82561.1 GDIP01218982 JAJ04420.1 GDIQ01210249 JAK41476.1 CAEY01000074 AAWZ02028191 GDIQ01254295 JAJ97429.1 GDIP01218981 JAJ04421.1 GDHF01029919 GDHF01014446 GDHF01007738 GDHF01004258 JAI22395.1 JAI37868.1 JAI44576.1 JAI48056.1 CVRI01000057 CRL02497.1 GDIP01253412 JAI69989.1 GDIQ01120229 JAL31497.1 AFYH01037266 AFYH01037267 AFYH01037268 AFYH01037269 AFYH01037270 GDIP01138138 JAL65576.1 GDIP01199072 JAJ24330.1 GDIP01230526 JAI92875.1 GDIQ01221184 JAK30541.1 GDIP01253370 JAI70031.1 GDIQ01124931 JAL26795.1 FR904563 CDQ66945.1 GDIP01199071 JAJ24331.1 AXCN02000801 GDIP01209724 JAJ13678.1 GDIP01086083 JAM17632.1 GDIQ01115631 GDIQ01115630 JAL36096.1 GDIP01186631 JAJ36771.1 GDIQ01236431 JAK15294.1 LJIG01016118 KRT81561.1 GDIP01163173 JAJ60229.1 KK758711 KFP44204.1 GDIP01184131 JAJ39271.1
OWR48986.1 KQ459585 KPI98287.1 KZ150088 PZC73742.1 ODYU01011849 SOQ57824.1 GGFM01003801 MBW24552.1 GGFK01004431 MBW37752.1 ADMH02000877 ETN64791.1 GFDL01009132 JAV25913.1 GGFM01003567 MBW24318.1 DS231854 EDS38062.1 GGFJ01003300 MBW52441.1 GGFJ01003301 MBW52442.1 GFDL01009247 JAV25798.1 GGFK01004550 MBW37871.1 CH477232 EAT46873.1 AAAB01008964 GBYB01003442 JAG73209.1 EAA12325.4 APCN01002480 GEZM01061486 JAV70438.1 JXUM01017001 KQ560472 KXJ82231.1 GDAI01001905 JAI15698.1 DS235855 EEB18984.1 NCKU01000159 RWS16828.1 KK113991 KFM61498.1 GGMR01014767 MBY27386.1 MH393923 AZV23876.1 GFXV01006234 MBW18039.1 GGMS01012908 MBY82111.1 IACF01000468 LAB66242.1 QCYY01004418 ROT61031.1 KK115903 KFM66326.1 NCKV01000613 RWS30165.1 RQTK01000929 RUS73496.1 ATLV01014737 KE524984 KFB39483.1 KX096891 ANQ68316.1 JH431832 GDIQ01169164 JAK82561.1 GDIP01218982 JAJ04420.1 GDIQ01210249 JAK41476.1 CAEY01000074 AAWZ02028191 GDIQ01254295 JAJ97429.1 GDIP01218981 JAJ04421.1 GDHF01029919 GDHF01014446 GDHF01007738 GDHF01004258 JAI22395.1 JAI37868.1 JAI44576.1 JAI48056.1 CVRI01000057 CRL02497.1 GDIP01253412 JAI69989.1 GDIQ01120229 JAL31497.1 AFYH01037266 AFYH01037267 AFYH01037268 AFYH01037269 AFYH01037270 GDIP01138138 JAL65576.1 GDIP01199072 JAJ24330.1 GDIP01230526 JAI92875.1 GDIQ01221184 JAK30541.1 GDIP01253370 JAI70031.1 GDIQ01124931 JAL26795.1 FR904563 CDQ66945.1 GDIP01199071 JAJ24331.1 AXCN02000801 GDIP01209724 JAJ13678.1 GDIP01086083 JAM17632.1 GDIQ01115631 GDIQ01115630 JAL36096.1 GDIP01186631 JAJ36771.1 GDIQ01236431 JAK15294.1 LJIG01016118 KRT81561.1 GDIP01163173 JAJ60229.1 KK758711 KFP44204.1 GDIP01184131 JAJ39271.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000000673
UP000069272
+ More
UP000002320 UP000192223 UP000008820 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000069940 UP000249989 UP000075903 UP000009046 UP000075900 UP000076408 UP000285301 UP000054359 UP000075885 UP000283509 UP000087266 UP000288716 UP000271974 UP000030765 UP000192224 UP000261540 UP000015104 UP000001646 UP000183832 UP000076420 UP000008672 UP000193380 UP000075886
UP000002320 UP000192223 UP000008820 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000069940 UP000249989 UP000075903 UP000009046 UP000075900 UP000076408 UP000285301 UP000054359 UP000075885 UP000283509 UP000087266 UP000288716 UP000271974 UP000030765 UP000192224 UP000261540 UP000015104 UP000001646 UP000183832 UP000076420 UP000008672 UP000193380 UP000075886
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
E9JEG6
A0A0N1ICV2
A0A212F5H5
A0A194PY77
A0A2W1BF90
A0A2H1WZL8
+ More
A0A2M3Z7Q7 A0A2M4AAJ3 W5JKX5 A0A182F616 A0A1Q3FED4 A0A2M3Z799 B0W7M7 A0A2M4BHK9 A0A2M4BHD0 A0A1W4WGV9 A0A1Q3FE57 A0A2M4AAQ2 Q17JG7 A0A182UAE0 A0A1S4GXS5 A0A182WUA0 A0A0C9QS22 A0A182L1R2 Q7Q3U0 A0A182HUW8 A0A182JR64 A0A1Y1LC16 A0A182GSK7 A0A182UYX4 A0A0K8TNI5 E0W028 A0A182RC80 A0A182Y019 A0A443RNN7 A0A087T8Q9 A0A2S2PEQ0 A0A3T0JLN2 A0A2H8TVU6 A0A2S2QWH0 A0A2P2HWW7 A0A182P4V7 A0A423S9X6 A0A087TMI7 A0A1S3LPT9 A0A443SRS7 A0A3S1B2K8 A0A084VND9 A0A1B1GE10 A0A1W4ZEJ7 A0A3B3RGD9 T1J3Z2 A0A1W4ZMW1 A0A1S3LQ11 A0A0P5M571 A0A1S3LQ57 A0A0P4Z3H5 A0A3B3RGH1 A0A1S3LQU2 A0A0P5INF4 A0A1W4ZEJ8 A0A1W4ZN17 T1KHA7 A0A3B3RG57 A0A1S3LPN4 H9GEN1 A0A0P5FRE2 A0A0P4Z9V0 A0A0K8VG98 A0A1S3LPT3 A0A1S3LQ52 A0A1J1IVD3 A0A0P4WPP2 A0A1S3LQ07 A0A1S3LQT7 A0A2C9JBH3 A0A0P5Q211 H2ZVB3 A0A0P5SLX2 A0A0P5BIH7 A0A0P4YA23 A0A182YTT5 A0A0P5HWQ5 A0A0P4WSA2 A0A0N8C166 A0A060WQQ5 A0A0P5ATI5 A0A182QQX4 A0A0P5AQW0 H2ZVB2 A0A0P5WI87 A0A0P5QDI6 A0A0P5BBX7 A0A0P5H8Z7 A0A0T6B2J4 A0A0P5DD02 A0A091KUC9 A0A0P5BJ72
A0A2M3Z7Q7 A0A2M4AAJ3 W5JKX5 A0A182F616 A0A1Q3FED4 A0A2M3Z799 B0W7M7 A0A2M4BHK9 A0A2M4BHD0 A0A1W4WGV9 A0A1Q3FE57 A0A2M4AAQ2 Q17JG7 A0A182UAE0 A0A1S4GXS5 A0A182WUA0 A0A0C9QS22 A0A182L1R2 Q7Q3U0 A0A182HUW8 A0A182JR64 A0A1Y1LC16 A0A182GSK7 A0A182UYX4 A0A0K8TNI5 E0W028 A0A182RC80 A0A182Y019 A0A443RNN7 A0A087T8Q9 A0A2S2PEQ0 A0A3T0JLN2 A0A2H8TVU6 A0A2S2QWH0 A0A2P2HWW7 A0A182P4V7 A0A423S9X6 A0A087TMI7 A0A1S3LPT9 A0A443SRS7 A0A3S1B2K8 A0A084VND9 A0A1B1GE10 A0A1W4ZEJ7 A0A3B3RGD9 T1J3Z2 A0A1W4ZMW1 A0A1S3LQ11 A0A0P5M571 A0A1S3LQ57 A0A0P4Z3H5 A0A3B3RGH1 A0A1S3LQU2 A0A0P5INF4 A0A1W4ZEJ8 A0A1W4ZN17 T1KHA7 A0A3B3RG57 A0A1S3LPN4 H9GEN1 A0A0P5FRE2 A0A0P4Z9V0 A0A0K8VG98 A0A1S3LPT3 A0A1S3LQ52 A0A1J1IVD3 A0A0P4WPP2 A0A1S3LQ07 A0A1S3LQT7 A0A2C9JBH3 A0A0P5Q211 H2ZVB3 A0A0P5SLX2 A0A0P5BIH7 A0A0P4YA23 A0A182YTT5 A0A0P5HWQ5 A0A0P4WSA2 A0A0N8C166 A0A060WQQ5 A0A0P5ATI5 A0A182QQX4 A0A0P5AQW0 H2ZVB2 A0A0P5WI87 A0A0P5QDI6 A0A0P5BBX7 A0A0P5H8Z7 A0A0T6B2J4 A0A0P5DD02 A0A091KUC9 A0A0P5BJ72
PDB
5FS7
E-value=1.32522e-68,
Score=660
Ontologies
KEGG
GO
PANTHER
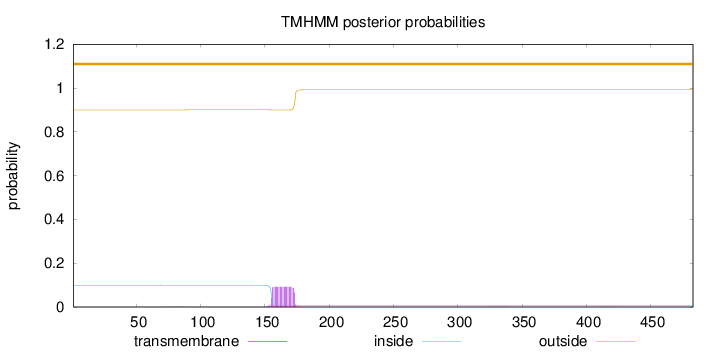
Topology
Length:
483
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.71726
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.09976
outside
1 - 483
Population Genetic Test Statistics
Pi
189.851526
Theta
152.826538
Tajima's D
1.354138
CLR
1.058822
CSRT
0.744612769361532
Interpretation
Uncertain
