Gene
KWMTBOMO08166
Pre Gene Modal
BGIBMGA014378
Annotation
lipase?_partial_[Bombyx_mori]
Full name
Lipase
+ More
Lipase 3
Lipase 3
Location in the cell
PlasmaMembrane Reliability : 1.406
Sequence
CDS
ATGTTGGCTGATGCCGGTTTCGACGTGTGGCTGGGTAATCTGAGAGGGAACTACTACACAGGTCACAGAACACTGAATAGAAGCGATCCAAGATTTTGGGAATTTAGCTTCGAAGAGCATGGTAAATACGACGCACCAGCCATGATAGACAAAGTCCTCAACGTGACAGGCCACCAGAAGATAATATACATAGGATATTCTATGGGAACAACCACCTTCTTCACGTTCATGTCCTCACATCCAGAATACAAAAGCAAAATCGTATCTTTCATTGCTCTGGCCCCTGCCGTATATCTGGACAACGTCAAACCACTGGCGACTATGTTTTTGAAGAATATGAACATACCCGACATAATGCGATCGAGAGGATTTATATCAGCAACATTCGAACCAGAATTCAAAAAGTTCTTAGGAACTTTATGCAACTTTAGGAACCCAAAGACTGATGTATGCTCGTCCCTTATAAACCTTGTAGTCGGAGAAGACAATGAACAGACCGATCCGGTATATGTTGGTGGCAAGAATGGAGCCGGCCTCTTGGAGACAGATCGAGCATTATGGGAAAATAGCTTTGACAGGTAA
Protein
MLADAGFDVWLGNLRGNYYTGHRTLNRSDPRFWEFSFEEHGKYDAPAMIDKVLNVTGHQKIIYIGYSMGTTTFFTFMSSHPEYKSKIVSFIALAPAVYLDNVKPLATMFLKNMNIPDIMRSRGFISATFEPEFKKFLGTLCNFRNPKTDVCSSLINLVVGEDNEQTDPVYVGGKNGAGLLETDRALWENSFDR
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Keywords
Complete proteome
Glycoprotein
Hydrolase
Lipid degradation
Lipid metabolism
Reference proteome
Signal
Feature
chain Lipase
Uniprot
H9JXW0
A0A0B6VL05
A0A194PY75
A0A2A4IVY6
A0A2W1BF38
A0A212F6V0
+ More
A0A194Q4F5 A0A2H1VIZ1 A0A0U4C5E4 A0A067R2Y8 A0A1E1XNF8 A0A182SDP3 A0A1B6G3S2 C8VV13 A0A0A1XQF1 O46108 A0A182VQ94 B4PQ03 A0A182MKD5 T1I6B5 T1GYY1 Q4V3V4 A0A1B6C389 A0A0M4ENG1 A0A1B6HSA1 B4HFU2 B4R1A4 A0A2P8XRN9 A0A232EX80 A0A1W4U7B6 B3P152 A0A2J7QW21 A0A226EG24 A0A1B6E4K7 E2AR09 E9IKU6 A0A182PA26 A0A1I7ZQM1 E2BWJ6 A0A182RCW3 A0A195AVW7 B4M6B9 B4NKC8 A0A232F2D1 A0A158P269 B4JI67 B3LX75 A0A3F2YX72 A0A2D4MFT8 K7J8S8 A0A1I7S4D9 A0A182H3N5 G3MH92 A0A182Y799 A0A0K8VD62 A0A0X3P504 A0A034VW56 A0A226E1Q5 A0A067R0F9 A0A183T3G1 B4KA89 A0A293MN23 A0A3P7P8G7 Q29A22 B4G3S6 A0A0X3PPU0 A0A2L2YLP4 A0A2A4JCC5 A0A131YJU9 A0A0N8DFT5 A0A0P6ANW7 A0A1I8NE89 A0A0P5ZUK5 A0A0P5AL58 A0A0P5UIW8 A0A1D1VTH2 A0A182KCP4 A0A2P8YGF9 A0A0P5XN98 A0A182GFS8 A0A182NS69 A0A183U260 A0A3P7F3G8 A0A0P4Z6G8 A0A0P6GD18 A0A0P5NNF9 N6UPZ0 V5I953 A0A0B2UP15 A0A0P6A9R6 A0A0K8W418 A0A3B0K5A1 A0A226E3U2 A0A0A1XMG0 A0A195FK23
A0A194Q4F5 A0A2H1VIZ1 A0A0U4C5E4 A0A067R2Y8 A0A1E1XNF8 A0A182SDP3 A0A1B6G3S2 C8VV13 A0A0A1XQF1 O46108 A0A182VQ94 B4PQ03 A0A182MKD5 T1I6B5 T1GYY1 Q4V3V4 A0A1B6C389 A0A0M4ENG1 A0A1B6HSA1 B4HFU2 B4R1A4 A0A2P8XRN9 A0A232EX80 A0A1W4U7B6 B3P152 A0A2J7QW21 A0A226EG24 A0A1B6E4K7 E2AR09 E9IKU6 A0A182PA26 A0A1I7ZQM1 E2BWJ6 A0A182RCW3 A0A195AVW7 B4M6B9 B4NKC8 A0A232F2D1 A0A158P269 B4JI67 B3LX75 A0A3F2YX72 A0A2D4MFT8 K7J8S8 A0A1I7S4D9 A0A182H3N5 G3MH92 A0A182Y799 A0A0K8VD62 A0A0X3P504 A0A034VW56 A0A226E1Q5 A0A067R0F9 A0A183T3G1 B4KA89 A0A293MN23 A0A3P7P8G7 Q29A22 B4G3S6 A0A0X3PPU0 A0A2L2YLP4 A0A2A4JCC5 A0A131YJU9 A0A0N8DFT5 A0A0P6ANW7 A0A1I8NE89 A0A0P5ZUK5 A0A0P5AL58 A0A0P5UIW8 A0A1D1VTH2 A0A182KCP4 A0A2P8YGF9 A0A0P5XN98 A0A182GFS8 A0A182NS69 A0A183U260 A0A3P7F3G8 A0A0P4Z6G8 A0A0P6GD18 A0A0P5NNF9 N6UPZ0 V5I953 A0A0B2UP15 A0A0P6A9R6 A0A0K8W418 A0A3B0K5A1 A0A226E3U2 A0A0A1XMG0 A0A195FK23
EC Number
3.1.1.-
Pubmed
EMBL
BABH01040389
AB983537
BAQ20442.1
KQ459585
KPI98291.1
NWSH01005934
+ More
PCG63829.1 KZ150088 PZC73729.1 AGBW02009965 OWR49465.1 KPI98290.1 ODYU01002832 SOQ40805.1 KU218686 ALX00066.1 KK852794 KDR16427.1 GFAA01002855 JAU00580.1 GECZ01012738 JAS57031.1 BT099759 ACV82458.1 GBXI01001534 JAD12758.1 Y14367 AE014297 BT023257 CM000160 EDW97230.1 AXCM01001786 ACPB03003261 CAQQ02051168 BT023252 AAY55668.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 CP012526 ALC46226.1 GECU01030185 JAS77521.1 CH480815 EDW42327.1 CM000364 EDX13081.1 PYGN01001466 PSN34671.1 NNAY01001790 OXU22918.1 CH954181 EDV49241.1 NEVH01009768 PNF32787.1 LNIX01000004 OXA56097.1 GEDC01004454 JAS32844.1 GL441846 EFN64155.1 GL764026 EFZ18725.1 GL451131 EFN79952.1 KQ976731 KYM76202.1 CH940652 EDW59195.1 CH964272 EDW84058.1 NNAY01001167 OXU24914.1 ADTU01007002 CH916369 EDV92948.1 CH902617 EDV41675.2 AXCN02000051 IACM01095948 LAB31888.1 JXUM01107701 KQ565165 KXJ71268.1 JO841243 AEO32860.1 GDHF01015819 JAI36495.1 GEEE01016161 JAP47064.1 GAKP01013224 JAC45728.1 LNIX01000009 OXA50396.1 KK852827 KDR15388.1 UYSU01036234 VDL97394.1 CH933806 EDW14576.1 GFWV01022365 GFWV01022368 GFWV01022370 MAA47097.1 UYRU01065522 VDN16352.1 CM000070 EAL27529.3 CH479179 EDW25031.1 GEEE01009270 JAP53955.1 IAAA01033384 LAA08380.1 NWSH01002126 PCG69082.1 GEDV01008984 JAP79573.1 GDIP01037967 JAM65748.1 GDIP01040265 JAM63450.1 GDIP01039254 JAM64461.1 GDIP01202418 JAJ20984.1 GDIP01114530 JAL89184.1 BDGG01000011 GAV04820.1 PYGN01000614 PSN43337.1 GDIP01072001 JAM31714.1 JXUM01060248 KQ562094 KXJ76705.1 UYWY01002680 VDM28293.1 GDIP01218767 JAJ04635.1 GDIQ01034971 JAN59766.1 GDIQ01139959 JAL11767.1 APGK01009375 APGK01009376 KB737932 ENN82831.1 GALX01003535 JAB64931.1 JPKZ01003222 KHN72716.1 GDIP01032305 JAM71410.1 GDHF01006507 JAI45807.1 OUUW01000013 SPP87872.1 LNIX01000007 OXA51571.1 GBXI01002237 JAD12055.1 KQ981523 KYN40354.1
PCG63829.1 KZ150088 PZC73729.1 AGBW02009965 OWR49465.1 KPI98290.1 ODYU01002832 SOQ40805.1 KU218686 ALX00066.1 KK852794 KDR16427.1 GFAA01002855 JAU00580.1 GECZ01012738 JAS57031.1 BT099759 ACV82458.1 GBXI01001534 JAD12758.1 Y14367 AE014297 BT023257 CM000160 EDW97230.1 AXCM01001786 ACPB03003261 CAQQ02051168 BT023252 AAY55668.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 CP012526 ALC46226.1 GECU01030185 JAS77521.1 CH480815 EDW42327.1 CM000364 EDX13081.1 PYGN01001466 PSN34671.1 NNAY01001790 OXU22918.1 CH954181 EDV49241.1 NEVH01009768 PNF32787.1 LNIX01000004 OXA56097.1 GEDC01004454 JAS32844.1 GL441846 EFN64155.1 GL764026 EFZ18725.1 GL451131 EFN79952.1 KQ976731 KYM76202.1 CH940652 EDW59195.1 CH964272 EDW84058.1 NNAY01001167 OXU24914.1 ADTU01007002 CH916369 EDV92948.1 CH902617 EDV41675.2 AXCN02000051 IACM01095948 LAB31888.1 JXUM01107701 KQ565165 KXJ71268.1 JO841243 AEO32860.1 GDHF01015819 JAI36495.1 GEEE01016161 JAP47064.1 GAKP01013224 JAC45728.1 LNIX01000009 OXA50396.1 KK852827 KDR15388.1 UYSU01036234 VDL97394.1 CH933806 EDW14576.1 GFWV01022365 GFWV01022368 GFWV01022370 MAA47097.1 UYRU01065522 VDN16352.1 CM000070 EAL27529.3 CH479179 EDW25031.1 GEEE01009270 JAP53955.1 IAAA01033384 LAA08380.1 NWSH01002126 PCG69082.1 GEDV01008984 JAP79573.1 GDIP01037967 JAM65748.1 GDIP01040265 JAM63450.1 GDIP01039254 JAM64461.1 GDIP01202418 JAJ20984.1 GDIP01114530 JAL89184.1 BDGG01000011 GAV04820.1 PYGN01000614 PSN43337.1 GDIP01072001 JAM31714.1 JXUM01060248 KQ562094 KXJ76705.1 UYWY01002680 VDM28293.1 GDIP01218767 JAJ04635.1 GDIQ01034971 JAN59766.1 GDIQ01139959 JAL11767.1 APGK01009375 APGK01009376 KB737932 ENN82831.1 GALX01003535 JAB64931.1 JPKZ01003222 KHN72716.1 GDIP01032305 JAM71410.1 GDHF01006507 JAI45807.1 OUUW01000013 SPP87872.1 LNIX01000007 OXA51571.1 GBXI01002237 JAD12055.1 KQ981523 KYN40354.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000027135
UP000075901
+ More
UP000000803 UP000075920 UP000002282 UP000075883 UP000015103 UP000015102 UP000092553 UP000001292 UP000000304 UP000245037 UP000215335 UP000192221 UP000008711 UP000235965 UP000198287 UP000000311 UP000075885 UP000095287 UP000008237 UP000075900 UP000078540 UP000008792 UP000007798 UP000005205 UP000001070 UP000007801 UP000075886 UP000002358 UP000095284 UP000069940 UP000249989 UP000076408 UP000050788 UP000275846 UP000009192 UP000001819 UP000008744 UP000095301 UP000186922 UP000075881 UP000075884 UP000050794 UP000267007 UP000019118 UP000031036 UP000268350 UP000078541
UP000000803 UP000075920 UP000002282 UP000075883 UP000015103 UP000015102 UP000092553 UP000001292 UP000000304 UP000245037 UP000215335 UP000192221 UP000008711 UP000235965 UP000198287 UP000000311 UP000075885 UP000095287 UP000008237 UP000075900 UP000078540 UP000008792 UP000007798 UP000005205 UP000001070 UP000007801 UP000075886 UP000002358 UP000095284 UP000069940 UP000249989 UP000076408 UP000050788 UP000275846 UP000009192 UP000001819 UP000008744 UP000095301 UP000186922 UP000075881 UP000075884 UP000050794 UP000267007 UP000019118 UP000031036 UP000268350 UP000078541
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JXW0
A0A0B6VL05
A0A194PY75
A0A2A4IVY6
A0A2W1BF38
A0A212F6V0
+ More
A0A194Q4F5 A0A2H1VIZ1 A0A0U4C5E4 A0A067R2Y8 A0A1E1XNF8 A0A182SDP3 A0A1B6G3S2 C8VV13 A0A0A1XQF1 O46108 A0A182VQ94 B4PQ03 A0A182MKD5 T1I6B5 T1GYY1 Q4V3V4 A0A1B6C389 A0A0M4ENG1 A0A1B6HSA1 B4HFU2 B4R1A4 A0A2P8XRN9 A0A232EX80 A0A1W4U7B6 B3P152 A0A2J7QW21 A0A226EG24 A0A1B6E4K7 E2AR09 E9IKU6 A0A182PA26 A0A1I7ZQM1 E2BWJ6 A0A182RCW3 A0A195AVW7 B4M6B9 B4NKC8 A0A232F2D1 A0A158P269 B4JI67 B3LX75 A0A3F2YX72 A0A2D4MFT8 K7J8S8 A0A1I7S4D9 A0A182H3N5 G3MH92 A0A182Y799 A0A0K8VD62 A0A0X3P504 A0A034VW56 A0A226E1Q5 A0A067R0F9 A0A183T3G1 B4KA89 A0A293MN23 A0A3P7P8G7 Q29A22 B4G3S6 A0A0X3PPU0 A0A2L2YLP4 A0A2A4JCC5 A0A131YJU9 A0A0N8DFT5 A0A0P6ANW7 A0A1I8NE89 A0A0P5ZUK5 A0A0P5AL58 A0A0P5UIW8 A0A1D1VTH2 A0A182KCP4 A0A2P8YGF9 A0A0P5XN98 A0A182GFS8 A0A182NS69 A0A183U260 A0A3P7F3G8 A0A0P4Z6G8 A0A0P6GD18 A0A0P5NNF9 N6UPZ0 V5I953 A0A0B2UP15 A0A0P6A9R6 A0A0K8W418 A0A3B0K5A1 A0A226E3U2 A0A0A1XMG0 A0A195FK23
A0A194Q4F5 A0A2H1VIZ1 A0A0U4C5E4 A0A067R2Y8 A0A1E1XNF8 A0A182SDP3 A0A1B6G3S2 C8VV13 A0A0A1XQF1 O46108 A0A182VQ94 B4PQ03 A0A182MKD5 T1I6B5 T1GYY1 Q4V3V4 A0A1B6C389 A0A0M4ENG1 A0A1B6HSA1 B4HFU2 B4R1A4 A0A2P8XRN9 A0A232EX80 A0A1W4U7B6 B3P152 A0A2J7QW21 A0A226EG24 A0A1B6E4K7 E2AR09 E9IKU6 A0A182PA26 A0A1I7ZQM1 E2BWJ6 A0A182RCW3 A0A195AVW7 B4M6B9 B4NKC8 A0A232F2D1 A0A158P269 B4JI67 B3LX75 A0A3F2YX72 A0A2D4MFT8 K7J8S8 A0A1I7S4D9 A0A182H3N5 G3MH92 A0A182Y799 A0A0K8VD62 A0A0X3P504 A0A034VW56 A0A226E1Q5 A0A067R0F9 A0A183T3G1 B4KA89 A0A293MN23 A0A3P7P8G7 Q29A22 B4G3S6 A0A0X3PPU0 A0A2L2YLP4 A0A2A4JCC5 A0A131YJU9 A0A0N8DFT5 A0A0P6ANW7 A0A1I8NE89 A0A0P5ZUK5 A0A0P5AL58 A0A0P5UIW8 A0A1D1VTH2 A0A182KCP4 A0A2P8YGF9 A0A0P5XN98 A0A182GFS8 A0A182NS69 A0A183U260 A0A3P7F3G8 A0A0P4Z6G8 A0A0P6GD18 A0A0P5NNF9 N6UPZ0 V5I953 A0A0B2UP15 A0A0P6A9R6 A0A0K8W418 A0A3B0K5A1 A0A226E3U2 A0A0A1XMG0 A0A195FK23
PDB
1K8Q
E-value=1.38614e-21,
Score=250
Ontologies
PATHWAY
PANTHER
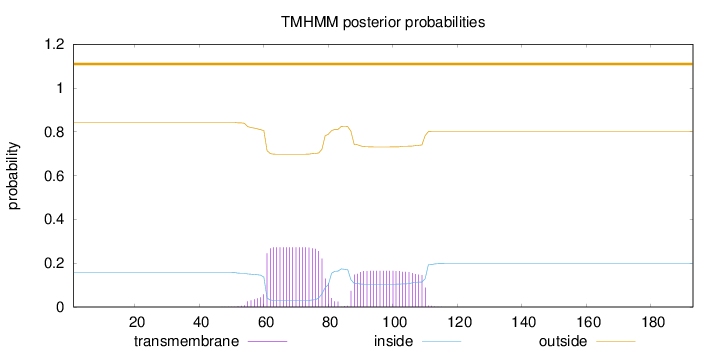
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.07300999999998
Exp number, first 60 AAs:
0.25027
Total prob of N-in:
0.15735
outside
1 - 193
Population Genetic Test Statistics
Pi
182.036213
Theta
177.836681
Tajima's D
0.067708
CLR
1.886436
CSRT
0.386830658467077
Interpretation
Uncertain
