Gene
KWMTBOMO08161
Annotation
putative_dynein?_axonemal?_heavy_polypeptide_1_[Danaus_plexippus]
Full name
SAGA-associated factor 11 homolog
Location in the cell
Cytoplasmic Reliability : 3.109
Sequence
CDS
ATGAAGGAGAGACACTGGAACGAGTTCATGGAGAAAGCCGGTATAACCCTGACCATGAACGAGAAGCAGACGTTCGACATGTGTCTCAAGCAAGGCGTCGGCAAACACGGCGACCTCATCGCGCAGATCGGTGAGCTCGCTTCCAAGGAGTACGTCATCGAGCAGGCCCTGGACAAGATGCAGACGGACTGGGCCACCAAGACCATGGAGATCACGCCTTACAAGAACACCGGAACGTACATGATGAAGGTCCCCGACGAGACCCTCCAGTTGTTGGACGAGCACCTGCTGGCCACCCAGCAACTCGGCTTCAGTCCGTTCAAAGCTGCCTTTGAACTCCGTATCGAGGAGTGGGACAGCAAGTTGAGGATGATGCAGATGGTTCTGGATGAATGGATCGAATGCCAGAAGGACTGGATGTACCTAGAGCCTATATTTACGTCGGAGGACATCTCCAGACAGCTCCCCTTCGAGGCGAAGAAGTACGGGACCATGGAGAGGATATGGAGGAGGATCATGGGGACGGCGGCCGCGTGTCCCAAGATGCTGGCGAGCGCGGAGGGCGCGGTGCTGGAGAGCGTGCGGGAGGCGCGCGGGCTGCTGGCGGCGGTGGCGCGCGGCCTGGCGCACTACACGGCGCGGAAGAGGCTCTGCTTCCCGCGCTTCTTCTTCCTCAGCGACGACGACCTGCTGCTCATCCTGGCGCACGCGCGGGACCCCGCCTCCGTCCAGCCGCATCTCAAGAAGTGTTTCGAGAATATTGCCAAGGTTCGTCTATCTTATCTTATATCTTTAAACGAGCAATTTTTGTATATATATAATATGAATCTCGGAAACGGCCCCAACGAGTTTCATAAGATTTAG
Protein
MKERHWNEFMEKAGITLTMNEKQTFDMCLKQGVGKHGDLIAQIGELASKEYVIEQALDKMQTDWATKTMEITPYKNTGTYMMKVPDETLQLLDEHLLATQQLGFSPFKAAFELRIEEWDSKLRMMQMVLDEWIECQKDWMYLEPIFTSEDISRQLPFEAKKYGTMERIWRRIMGTAAACPKMLASAEGAVLESVREARGLLAAVARGLAHYTARKRLCFPRFFFLSDDDLLLILAHARDPASVQPHLKKCFENIAKVRLSYLISLNEQFLYIYNMNLGNGPNEFHKI
Summary
Description
Component of the transcription regulatory histone acetylation (HAT) complex SAGA, a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates histone H2B. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription.
Subunit
Component of some SAGA transcription coactivator-HAT complexes. Within the SAGA complex, participates to a subcomplex of SAGA called the DUB module (deubiquitination module).
Similarity
Belongs to the SGF11 family.
Uniprot
A0A2A4JDQ9
A0A2A4JD05
A0A2A4JE46
A0A2H1VXK0
H9JXU6
A0A3S2M4I0
+ More
A0A1B6E076 A0A1B6CKQ5 A0A1B6F3E5 A0A2P8YP94 A7S0X9 E0VBG0 H3BFH9 A0A0L8HDN8 A0A2B4S5H5 A0A3M6UN98 D2A3I1 W4ZJK2 A0A2G8K162 W4ZLH1 A0A1W4X6Z1 A0A3B3QM65 A0A3S0ZN14 A0A1Y1N5M7 V3ZE33 A0A2T7P6N6 A0A1S3LD14 A0A1S3LD11 A0A3B4E2P3 A0A369RVY1 B3SBZ0 A0A2R2MMP5 A0A2C9JK78 A0A2R2MT18 A0A2C9JK70 A0A060XHS4 A0A183KTH7 A0A419PK76 G7YJ50 A0A267D9H9 A0A1I8GJ57 K1RJM8 A0A1I8HE72 U4UG54 A0A2T7P6M7 A0A183LAJ4 A0A3P8XPP2 A0A2D0PK34 A0A2D0PK64 A0A2D0PIM8 A0A074ZNX3 A0A1S8WJX6 A0A1L8GPB6 H2YF94 R7VGC6 A0A1X7U4E6 A0A3Q3E168 A0A0R4IHI4 A0A0R4IY79 A0A3N0XG01 A0A2U9BH60 A0A401SWB1 A0A095A169 F6PIN6 A0A430QF48 A0A183NRQ5 W5NAX8 H0X626 A0A177BAD3 I3MF05 A0A1W4Z8M3 A0A1W4Z8M5 M3YZ26 A0A0X3P6T8 A0A183A8Q8 A0A1A6G290 A0A183T112 A0A0X3NTI6 A0A0X3QBN0 A0A0V0J7D3 F6XWP3 G3N1C8 A0A2K6RAT4 A0A3Q3STI9 A0A402FM20 A0A2Y9IYH5 H0V0D2 A0A2K6RAQ5 A0A3Q2HHT5 A0A3B4VGS0 A0A1S3GBV6 A0A2K6MR81 A0A3Q0DG64 A0A3Q4IDQ3 L8IEV2 A0A2U3XK15 A0A452EGB0 G3TQL9 A0A0R3TYZ3
A0A1B6E076 A0A1B6CKQ5 A0A1B6F3E5 A0A2P8YP94 A7S0X9 E0VBG0 H3BFH9 A0A0L8HDN8 A0A2B4S5H5 A0A3M6UN98 D2A3I1 W4ZJK2 A0A2G8K162 W4ZLH1 A0A1W4X6Z1 A0A3B3QM65 A0A3S0ZN14 A0A1Y1N5M7 V3ZE33 A0A2T7P6N6 A0A1S3LD14 A0A1S3LD11 A0A3B4E2P3 A0A369RVY1 B3SBZ0 A0A2R2MMP5 A0A2C9JK78 A0A2R2MT18 A0A2C9JK70 A0A060XHS4 A0A183KTH7 A0A419PK76 G7YJ50 A0A267D9H9 A0A1I8GJ57 K1RJM8 A0A1I8HE72 U4UG54 A0A2T7P6M7 A0A183LAJ4 A0A3P8XPP2 A0A2D0PK34 A0A2D0PK64 A0A2D0PIM8 A0A074ZNX3 A0A1S8WJX6 A0A1L8GPB6 H2YF94 R7VGC6 A0A1X7U4E6 A0A3Q3E168 A0A0R4IHI4 A0A0R4IY79 A0A3N0XG01 A0A2U9BH60 A0A401SWB1 A0A095A169 F6PIN6 A0A430QF48 A0A183NRQ5 W5NAX8 H0X626 A0A177BAD3 I3MF05 A0A1W4Z8M3 A0A1W4Z8M5 M3YZ26 A0A0X3P6T8 A0A183A8Q8 A0A1A6G290 A0A183T112 A0A0X3NTI6 A0A0X3QBN0 A0A0V0J7D3 F6XWP3 G3N1C8 A0A2K6RAT4 A0A3Q3STI9 A0A402FM20 A0A2Y9IYH5 H0V0D2 A0A2K6RAQ5 A0A3Q2HHT5 A0A3B4VGS0 A0A1S3GBV6 A0A2K6MR81 A0A3Q0DG64 A0A3Q4IDQ3 L8IEV2 A0A2U3XK15 A0A452EGB0 G3TQL9 A0A0R3TYZ3
Pubmed
19121390
29403074
17615350
20566863
9215903
30382153
+ More
18362917 19820115 29023486 29240929 28004739 23254933 30042472 18719581 15562597 24755649 22023798 26392545 22992520 23537049 25069045 27762356 23594743 30297745 22246508 20431018 17495919 19393038 25362486 21993624 19892987 26319212 25186727 22751099
18362917 19820115 29023486 29240929 28004739 23254933 30042472 18719581 15562597 24755649 22023798 26392545 22992520 23537049 25069045 27762356 23594743 30297745 22246508 20431018 17495919 19393038 25362486 21993624 19892987 26319212 25186727 22751099
EMBL
NWSH01001830
PCG69979.1
PCG69977.1
PCG69978.1
ODYU01005066
SOQ45577.1
+ More
BABH01040386 BABH01040387 RSAL01000037 RVE51194.1 GEDC01005987 JAS31311.1 GEDC01023330 JAS13968.1 GECZ01025168 JAS44601.1 PYGN01000453 PSN46069.1 DS469562 EDO42618.1 DS235030 EEB10716.1 AFYH01005848 AFYH01005849 AFYH01005850 AFYH01005851 AFYH01005852 AFYH01005853 AFYH01005854 AFYH01005855 AFYH01005856 AFYH01005857 KQ418416 KOF87423.1 LSMT01000171 PFX24646.1 RCHS01001119 RMX55115.1 KQ971338 EFA01906.2 AAGJ04100766 AAGJ04100767 AAGJ04100768 AAGJ04100769 AAGJ04100770 AAGJ04100771 MRZV01000992 PIK41736.1 AAGJ04100772 AAGJ04100773 RQTK01000491 RUS78759.1 GEZM01017151 GEZM01017145 JAV90907.1 KB202619 ESO89348.1 PZQS01000006 PVD29061.1 NOWV01000159 RDD38836.1 DS985267 EDV19716.1 FR905415 CDQ79158.1 UZAK01040924 VDP65585.1 NIRI01002318 RJW62475.1 DF143394 GAA52983.1 NIVC01005167 PAA45958.1 JH816168 EKC41830.1 KB632308 ERL92012.1 PVD29059.1 UZAI01000160 VDO49358.1 KL596691 KER28811.1 KV906440 OON14744.1 CM004472 OCT85694.1 AMQN01004546 AMQN01004547 KB293809 ELU15361.1 FO704861 FQ311885 RJVU01075544 ROI16260.1 CP026248 AWP03287.1 BEZZ01000623 GCC34692.1 KL251154 KGB39209.1 AAMC01048030 AAMC01048031 AAMC01048032 AAMC01048033 QMKO01001855 RTG86126.1 UZAL01026724 VDP25180.1 AHAT01014842 AHAT01014843 AHAT01014844 AAQR03061720 AAQR03061721 AAQR03061722 AAQR03061723 AAQR03061724 AAQR03061725 AAQR03061726 AAQR03061727 LWCA01000141 OAF70501.1 AGTP01005611 AGTP01005612 AEYP01001932 GEEE01015646 JAP47579.1 UZAN01040308 VDP69207.1 LZPO01107936 OBS59920.1 UYSU01035690 VDL96547.1 GEEE01020327 JAP42898.1 GEEE01006499 JAP56726.1 GEEE01002213 JAP61012.1 BDOT01000329 GCF57219.1 AAKN02001401 AAKN02001402 JH881365 ELR54718.1 LWLT01000024 UZAE01014939 VDO14848.1
BABH01040386 BABH01040387 RSAL01000037 RVE51194.1 GEDC01005987 JAS31311.1 GEDC01023330 JAS13968.1 GECZ01025168 JAS44601.1 PYGN01000453 PSN46069.1 DS469562 EDO42618.1 DS235030 EEB10716.1 AFYH01005848 AFYH01005849 AFYH01005850 AFYH01005851 AFYH01005852 AFYH01005853 AFYH01005854 AFYH01005855 AFYH01005856 AFYH01005857 KQ418416 KOF87423.1 LSMT01000171 PFX24646.1 RCHS01001119 RMX55115.1 KQ971338 EFA01906.2 AAGJ04100766 AAGJ04100767 AAGJ04100768 AAGJ04100769 AAGJ04100770 AAGJ04100771 MRZV01000992 PIK41736.1 AAGJ04100772 AAGJ04100773 RQTK01000491 RUS78759.1 GEZM01017151 GEZM01017145 JAV90907.1 KB202619 ESO89348.1 PZQS01000006 PVD29061.1 NOWV01000159 RDD38836.1 DS985267 EDV19716.1 FR905415 CDQ79158.1 UZAK01040924 VDP65585.1 NIRI01002318 RJW62475.1 DF143394 GAA52983.1 NIVC01005167 PAA45958.1 JH816168 EKC41830.1 KB632308 ERL92012.1 PVD29059.1 UZAI01000160 VDO49358.1 KL596691 KER28811.1 KV906440 OON14744.1 CM004472 OCT85694.1 AMQN01004546 AMQN01004547 KB293809 ELU15361.1 FO704861 FQ311885 RJVU01075544 ROI16260.1 CP026248 AWP03287.1 BEZZ01000623 GCC34692.1 KL251154 KGB39209.1 AAMC01048030 AAMC01048031 AAMC01048032 AAMC01048033 QMKO01001855 RTG86126.1 UZAL01026724 VDP25180.1 AHAT01014842 AHAT01014843 AHAT01014844 AAQR03061720 AAQR03061721 AAQR03061722 AAQR03061723 AAQR03061724 AAQR03061725 AAQR03061726 AAQR03061727 LWCA01000141 OAF70501.1 AGTP01005611 AGTP01005612 AEYP01001932 GEEE01015646 JAP47579.1 UZAN01040308 VDP69207.1 LZPO01107936 OBS59920.1 UYSU01035690 VDL96547.1 GEEE01020327 JAP42898.1 GEEE01006499 JAP56726.1 GEEE01002213 JAP61012.1 BDOT01000329 GCF57219.1 AAKN02001401 AAKN02001402 JH881365 ELR54718.1 LWLT01000024 UZAE01014939 VDO14848.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000245037
UP000001593
UP000009046
+ More
UP000008672 UP000053454 UP000225706 UP000275408 UP000007266 UP000007110 UP000230750 UP000192223 UP000261540 UP000271974 UP000030746 UP000245119 UP000087266 UP000261440 UP000253843 UP000009022 UP000085678 UP000076420 UP000193380 UP000050789 UP000215902 UP000095280 UP000005408 UP000030742 UP000050790 UP000277204 UP000265140 UP000221080 UP000186698 UP000007875 UP000014760 UP000007879 UP000261660 UP000000437 UP000246464 UP000287033 UP000008143 UP000050791 UP000018468 UP000005225 UP000005215 UP000192224 UP000000715 UP000050740 UP000092124 UP000050788 UP000275846 UP000002280 UP000009136 UP000233200 UP000261640 UP000288954 UP000248482 UP000005447 UP000002281 UP000261420 UP000081671 UP000233180 UP000189706 UP000261580 UP000245341 UP000291000 UP000007646 UP000046398 UP000278807
UP000008672 UP000053454 UP000225706 UP000275408 UP000007266 UP000007110 UP000230750 UP000192223 UP000261540 UP000271974 UP000030746 UP000245119 UP000087266 UP000261440 UP000253843 UP000009022 UP000085678 UP000076420 UP000193380 UP000050789 UP000215902 UP000095280 UP000005408 UP000030742 UP000050790 UP000277204 UP000265140 UP000221080 UP000186698 UP000007875 UP000014760 UP000007879 UP000261660 UP000000437 UP000246464 UP000287033 UP000008143 UP000050791 UP000018468 UP000005225 UP000005215 UP000192224 UP000000715 UP000050740 UP000092124 UP000050788 UP000275846 UP000002280 UP000009136 UP000233200 UP000261640 UP000288954 UP000248482 UP000005447 UP000002281 UP000261420 UP000081671 UP000233180 UP000189706 UP000261580 UP000245341 UP000291000 UP000007646 UP000046398 UP000278807
PRIDE
Pfam
Interpro
IPR042228
Dynein_2_C
+ More
IPR035699 AAA_6
IPR013602 Dynein_heavy_dom-2
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR026983 DHC_fam
IPR003593 AAA+_ATPase
IPR026975 DNAH1
IPR024317 Dynein_heavy_chain_D4_dom
IPR041589 DNAH3_AAA_lid_1
IPR004273 Dynein_heavy_D6_P-loop
IPR042219 AAA_lid_11_sf
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR035706 AAA_9
IPR024743 Dynein_HC_stalk
IPR013246 SAGA_su_Sgf11
IPR013243 SCA7_dom
IPR016024 ARM-type_fold
IPR035699 AAA_6
IPR013602 Dynein_heavy_dom-2
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR026983 DHC_fam
IPR003593 AAA+_ATPase
IPR026975 DNAH1
IPR024317 Dynein_heavy_chain_D4_dom
IPR041589 DNAH3_AAA_lid_1
IPR004273 Dynein_heavy_D6_P-loop
IPR042219 AAA_lid_11_sf
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR035706 AAA_9
IPR024743 Dynein_HC_stalk
IPR013246 SAGA_su_Sgf11
IPR013243 SCA7_dom
IPR016024 ARM-type_fold
Gene 3D
ProteinModelPortal
A0A2A4JDQ9
A0A2A4JD05
A0A2A4JE46
A0A2H1VXK0
H9JXU6
A0A3S2M4I0
+ More
A0A1B6E076 A0A1B6CKQ5 A0A1B6F3E5 A0A2P8YP94 A7S0X9 E0VBG0 H3BFH9 A0A0L8HDN8 A0A2B4S5H5 A0A3M6UN98 D2A3I1 W4ZJK2 A0A2G8K162 W4ZLH1 A0A1W4X6Z1 A0A3B3QM65 A0A3S0ZN14 A0A1Y1N5M7 V3ZE33 A0A2T7P6N6 A0A1S3LD14 A0A1S3LD11 A0A3B4E2P3 A0A369RVY1 B3SBZ0 A0A2R2MMP5 A0A2C9JK78 A0A2R2MT18 A0A2C9JK70 A0A060XHS4 A0A183KTH7 A0A419PK76 G7YJ50 A0A267D9H9 A0A1I8GJ57 K1RJM8 A0A1I8HE72 U4UG54 A0A2T7P6M7 A0A183LAJ4 A0A3P8XPP2 A0A2D0PK34 A0A2D0PK64 A0A2D0PIM8 A0A074ZNX3 A0A1S8WJX6 A0A1L8GPB6 H2YF94 R7VGC6 A0A1X7U4E6 A0A3Q3E168 A0A0R4IHI4 A0A0R4IY79 A0A3N0XG01 A0A2U9BH60 A0A401SWB1 A0A095A169 F6PIN6 A0A430QF48 A0A183NRQ5 W5NAX8 H0X626 A0A177BAD3 I3MF05 A0A1W4Z8M3 A0A1W4Z8M5 M3YZ26 A0A0X3P6T8 A0A183A8Q8 A0A1A6G290 A0A183T112 A0A0X3NTI6 A0A0X3QBN0 A0A0V0J7D3 F6XWP3 G3N1C8 A0A2K6RAT4 A0A3Q3STI9 A0A402FM20 A0A2Y9IYH5 H0V0D2 A0A2K6RAQ5 A0A3Q2HHT5 A0A3B4VGS0 A0A1S3GBV6 A0A2K6MR81 A0A3Q0DG64 A0A3Q4IDQ3 L8IEV2 A0A2U3XK15 A0A452EGB0 G3TQL9 A0A0R3TYZ3
A0A1B6E076 A0A1B6CKQ5 A0A1B6F3E5 A0A2P8YP94 A7S0X9 E0VBG0 H3BFH9 A0A0L8HDN8 A0A2B4S5H5 A0A3M6UN98 D2A3I1 W4ZJK2 A0A2G8K162 W4ZLH1 A0A1W4X6Z1 A0A3B3QM65 A0A3S0ZN14 A0A1Y1N5M7 V3ZE33 A0A2T7P6N6 A0A1S3LD14 A0A1S3LD11 A0A3B4E2P3 A0A369RVY1 B3SBZ0 A0A2R2MMP5 A0A2C9JK78 A0A2R2MT18 A0A2C9JK70 A0A060XHS4 A0A183KTH7 A0A419PK76 G7YJ50 A0A267D9H9 A0A1I8GJ57 K1RJM8 A0A1I8HE72 U4UG54 A0A2T7P6M7 A0A183LAJ4 A0A3P8XPP2 A0A2D0PK34 A0A2D0PK64 A0A2D0PIM8 A0A074ZNX3 A0A1S8WJX6 A0A1L8GPB6 H2YF94 R7VGC6 A0A1X7U4E6 A0A3Q3E168 A0A0R4IHI4 A0A0R4IY79 A0A3N0XG01 A0A2U9BH60 A0A401SWB1 A0A095A169 F6PIN6 A0A430QF48 A0A183NRQ5 W5NAX8 H0X626 A0A177BAD3 I3MF05 A0A1W4Z8M3 A0A1W4Z8M5 M3YZ26 A0A0X3P6T8 A0A183A8Q8 A0A1A6G290 A0A183T112 A0A0X3NTI6 A0A0X3QBN0 A0A0V0J7D3 F6XWP3 G3N1C8 A0A2K6RAT4 A0A3Q3STI9 A0A402FM20 A0A2Y9IYH5 H0V0D2 A0A2K6RAQ5 A0A3Q2HHT5 A0A3B4VGS0 A0A1S3GBV6 A0A2K6MR81 A0A3Q0DG64 A0A3Q4IDQ3 L8IEV2 A0A2U3XK15 A0A452EGB0 G3TQL9 A0A0R3TYZ3
PDB
5NUG
E-value=3.73705e-19,
Score=231
Ontologies
GO
PANTHER
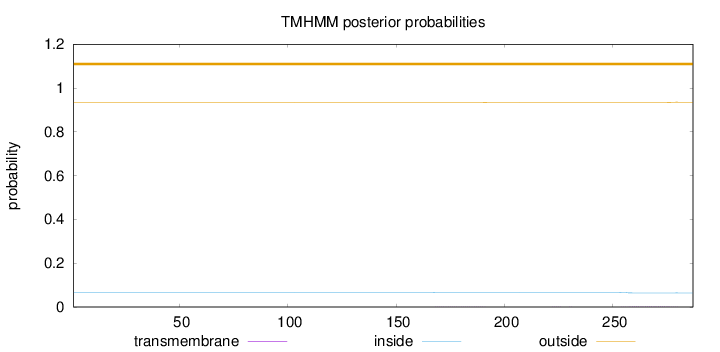
Topology
Subcellular location
Nucleus
Length:
287
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06173
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06671
outside
1 - 287
Population Genetic Test Statistics
Pi
323.368542
Theta
213.24667
Tajima's D
1.636347
CLR
0.014482
CSRT
0.817909104544773
Interpretation
Uncertain
