Gene
KWMTBOMO08159
Pre Gene Modal
BGIBMGA014367
Annotation
PREDICTED:_dynein_heavy_chain_1?_axonemal-like_[Papilio_polytes]
Full name
SAGA-associated factor 11 homolog
Location in the cell
Cytoplasmic Reliability : 1.697 PlasmaMembrane Reliability : 1.023
Sequence
CDS
ATGGCGGAAGTGTCCCTGCCCCACACCCCCGGAGCCACGCCCCTCTCCATCCTGGGGGACCCGGTGCAGATCAGGACGTGGCAGATGTACGGGCTGCCGCGGGACCCGCTGTCGGTGGAGTCCGCCGTGCTGATGGCGGCGTCGCGGCGCTGGCCCCTCGTCATCGACCCCCAGACGCAGGCCAACAACTGGATCCGAGCCATGGGAAAAATCGAAGGTCTGATAGTGTGCAAGCCGAACGACGCGGACCTGCTGCGCAGCTTCGAGTCCGCGCTGCGCTTCGGGAAGCCGCTGCTGCTGGAGAACGTGGGCCAGGAGCTGGACCCCGCGCTCGATCCCGTCCTCAAGCGTCAGTACTTCCGGCAAGCTGGGCAACTGGTGCTGAAGCTGGGCGAGTCGCTGATACCGTACGCGGCTGGCTTCCGCCTGTACATGACTACGAAGCTGCCTAACCCTGTGTACTCCCCGGACACGTCGGTCAAGGTGCAGGTCGTGAACTTCGCCCTGGTGCCCAGTGGGCTGTCGGAGCAGCTGCTGTCGGTGGTGGTGGCGCAGGAGCGGCCGGCGCTGGAGGAGACCCGCGGCGCGCTGGTGGTGTCGCGCGCGCAGCTGTTCGCGCAGCTGGCCACTATGGGCGCGCGCGTGCTGCACGGGCTGGCCTCCTGCCAGGCGCCCGTCGACGACCTGCAGCTCATCCTCACGCTCGAGGCCATCAAGATCAAGACGGCGGAGATCCTGACGAAGGTGTCGGACATGGAGCGCACCACAGCGGAGGTGGACGCCACGCGCGGCGAGTACACCGGCGTGGCGGCGCGCGGACAGATCCTGTTCTTCTGCGTGGCCGCGCTGGCCGCCGTGGACCCCATGTACCAGTACTCGCTCGACTGGTTCACCGGACTCTTCCTCAAGTCCATGGCGGAGACCGAGCCGAGCGAAGACATCCTGGAGCGCGTGGAGACGATCATCGAGCACTTCACGTTCTCGCTGTACCTGAACGTGTGCCGCAGCCTGTTCGAGCGACACAAGCTGCTGTTCGCCTTCCTGCTCACCGCCAGGATCCTGCTCGACGCAGGCGTCATCAGGACCCACGAATACAACTTCTTGATTAATGGTGCTTCATCCCAAGAAGCTTCCGTACCCGAAGGCACTGGACTCACAGCTGGACGCCTTCCAGAAGATCCTTGTGTTGAAGTGTATCCGGCCCGACAAAATGATCCCGGGTCTGCAGCGTGTCGGATGGTGCAGGAATACGTGTCGGCGGCCCGGGGGGCGCGGTACGTGGAGGCGCGGGGCGGGGGAGGGCGCGCCGGGCTGGCCGCGCTGCTGGCCGACTCCTCCCCGCGCGCCCCCCTCGTGCTGGTGCTGTCCTCGGGCACTGACCCGGCCGCCGATTTGATGCAGTTCGCTGACAAGATGAAGATGGGCAAACGGTTCGAGAGCATCTCCCTGGGCCAGGGTCAGGGCCCCATCGCGGAAAACATGATCAGACTGGGCTGCGACTTTGGCAACTGGGTCTTCCTGCAGAACTGTCACCTGTCGCCCTCGTGGATGCCAGCGCTGGAGCTCCTGGTGGAACAACTCCAGGCTGAGAATGTGCACAAAGACTTCCGCCTGTGGCTCACCTCCACACCCTCGCCGCATTTCCCGGTATCGTTATTGCAGAATGGATACAAGATGACGATAGAACCTCCGCGGGGGGTGAAAGCGAACCTGCTGAAGGCTTACACGAACCAGATCCCGGACTTCGCAGACTTCTTTGAGTCGGATGAACCTAAAGTTCCCCAATTTAAGGTTTACTAA
Protein
MAEVSLPHTPGATPLSILGDPVQIRTWQMYGLPRDPLSVESAVLMAASRRWPLVIDPQTQANNWIRAMGKIEGLIVCKPNDADLLRSFESALRFGKPLLLENVGQELDPALDPVLKRQYFRQAGQLVLKLGESLIPYAAGFRLYMTTKLPNPVYSPDTSVKVQVVNFALVPSGLSEQLLSVVVAQERPALEETRGALVVSRAQLFAQLATMGARVLHGLASCQAPVDDLQLILTLEAIKIKTAEILTKVSDMERTTAEVDATRGEYTGVAARGQILFFCVAALAAVDPMYQYSLDWFTGLFLKSMAETEPSEDILERVETIIEHFTFSLYLNVCRSLFERHKLLFAFLLTARILLDAGVIRTHEYNFLINGASSQEASVPEGTGLTAGRLPEDPCVEVYPARQNDPGSAACRMVQEYVSAARGARYVEARGGGGRAGLAALLADSSPRAPLVLVLSSGTDPAADLMQFADKMKMGKRFESISLGQGQGPIAENMIRLGCDFGNWVFLQNCHLSPSWMPALELLVEQLQAENVHKDFRLWLTSTPSPHFPVSLLQNGYKMTIEPPRGVKANLLKAYTNQIPDFADFFESDEPKVPQFKVY
Summary
Description
Component of the transcription regulatory histone acetylation (HAT) complex SAGA, a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates histone H2B. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription.
Subunit
Component of some SAGA transcription coactivator-HAT complexes. Within the SAGA complex, participates to a subcomplex of SAGA called the DUB module (deubiquitination module).
Similarity
Belongs to the SGF11 family.
Uniprot
EMBL
AEFK01097765
KB202619
ESO89348.1
AMQN01004546
AMQN01004547
KB293809
+ More
ELU15361.1 NEDP02004067 OWF46845.1 JH816168 EKC41830.1 KQ042005 KKF19598.1 CABG01000035 CCD59439.1 RQTK01000491 RUS78759.1 KQ257450 KND05191.1 AGCU01056634 AGCU01056635 AGCU01056636 AGCU01056637 AGCU01056638 AGCU01056639 AGCU01056640 AGCU01056641 AGCU01056642 AGCU01056643
ELU15361.1 NEDP02004067 OWF46845.1 JH816168 EKC41830.1 KQ042005 KKF19598.1 CABG01000035 CCD59439.1 RQTK01000491 RUS78759.1 KQ257450 KND05191.1 AGCU01056634 AGCU01056635 AGCU01056636 AGCU01056637 AGCU01056638 AGCU01056639 AGCU01056640 AGCU01056641 AGCU01056642 AGCU01056643
Proteomes
Pfam
Interpro
IPR041228
Dynein_C
+ More
IPR041658 AAA_lid_11
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR004273 Dynein_heavy_D6_P-loop
IPR026975 DNAH1
IPR035706 AAA_9
IPR024743 Dynein_HC_stalk
IPR041466 Dynein_AAA5_ext
IPR013602 Dynein_heavy_dom-2
IPR035699 AAA_6
IPR042228 Dynein_2_C
IPR042222 Dynein_2_N
IPR041589 DNAH3_AAA_lid_1
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR013246 SAGA_su_Sgf11
IPR013243 SCA7_dom
IPR041658 AAA_lid_11
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR004273 Dynein_heavy_D6_P-loop
IPR026975 DNAH1
IPR035706 AAA_9
IPR024743 Dynein_HC_stalk
IPR041466 Dynein_AAA5_ext
IPR013602 Dynein_heavy_dom-2
IPR035699 AAA_6
IPR042228 Dynein_2_C
IPR042222 Dynein_2_N
IPR041589 DNAH3_AAA_lid_1
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR013246 SAGA_su_Sgf11
IPR013243 SCA7_dom
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
PDB
4RH7
E-value=3.73154e-48,
Score=485
Ontologies
GO
PANTHER
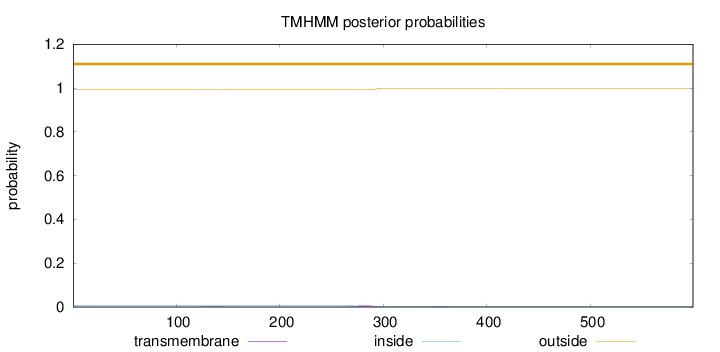
Topology
Subcellular location
Nucleus
Length:
599
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.227170000000001
Exp number, first 60 AAs:
0.00111
Total prob of N-in:
0.00795
outside
1 - 599
Population Genetic Test Statistics
Pi
203.932013
Theta
191.459957
Tajima's D
0.87438
CLR
0.02152
CSRT
0.625868706564672
Interpretation
Uncertain
