Gene
KWMTBOMO08154
Pre Gene Modal
BGIBMGA014369
Annotation
PREDICTED:_lysosomal_acid_phosphatase-like_[Amyelois_transitella]
Full name
Prostatic acid phosphatase
Alternative Name
5'-nucleotidase
Ecto-5'-nucleotidase
Fluoride-resistant acid phosphatase
Thiamine monophosphatase
Ecto-5'-nucleotidase
Fluoride-resistant acid phosphatase
Thiamine monophosphatase
Location in the cell
PlasmaMembrane Reliability : 0.898
Sequence
CDS
ATGCGAGTCGTGTACTTTGTAGGCGTCTTGGCTGCGTTGGCCCAGCACGTCAGGGCTGGCGACGCTACAGACGGAACCGAACTGGTTTCAACTTTTTTGGTACATCGGCACGGCGACAGGACGCCGGTGGAGATCACCCTGGCCATGTCCACGGACCCCGCCGCGCTGGAGCAGGCCGCGGCTCCCTATGGCTTTGGACAACTCACTGAGTTCGGTAAGAAGACCAGCTACAAAGTGGGAGAGTACATCAAGTCACGTTACGGCGCTCTGCTCTCGGACCGATACAACGCCTCCGAGATCTTCATCCGGTCCACGGACTCGACACGCACGAAGATGACTGTCCTGACCGCGATGGCCGCTGTCTACCCACCTCCTGAAGACAACTGGAGCTCAGAGATTCACTGGACCCCCGTGCCCTACACCACCGTCCCACCTAAATATGACCATAACCTGGCAAACCTCAACTGTCCGGAGTTCATCTCGTTCTACTACGACCAGTTCATGAAACCTTCCGAGAAGATGGCCCCCTACCGCGACTTCCTGGCGAGATGGTCTGAAATTCTCGGTTTCGACCTCGTGCAGATGCCCGCCCTGGCGTACATGTTCTACGACGTATACACTGCTCAGCTCGGGCTGGGCGTCCCGCTGGACGAGGCCACGGCGCAGAAGTTCCCCGAGATCGAAGCGGCGGCCGGGGCGGCCATCGACGTCGTGTTCGGAGATGAAAACTACATCGCGCTACAAGCGGGAGTGTTGCTGAATGAGTTCTTGAAGGCGGCGGCGGCGGTGGTGGGGGGCGAGAGCGCGCGGCGGGTGCACGTGTACTCCGCGCACGACTACAACGTGTACTCCCTGATGGCCGTCTCCAGGATCAGTCCGCGACAAGGAGTGCCCAAGTACGGCTCCGTATTCTCCCTCGAGCTGAGGAAGGTCACCGCGACCGGGAAGTATGTAGTTTTGCCCGTTTATCTAAGATCTCCGTACGAGCAGATCCAGTACCTGCAGGTGGAGGGCTGCAGCGCGCTGCTGTGTGACCTGGAAGAGTTTCAGCAGCTCACCTCCCGCTACGTGCTCGAAGAGGACGAGTGGAGGACGAAGTGCGGCTTCACGCACGACCTCGAAGTGGACGACTCCAGCATTAACTAA
Protein
MRVVYFVGVLAALAQHVRAGDATDGTELVSTFLVHRHGDRTPVEITLAMSTDPAALEQAAAPYGFGQLTEFGKKTSYKVGEYIKSRYGALLSDRYNASEIFIRSTDSTRTKMTVLTAMAAVYPPPEDNWSSEIHWTPVPYTTVPPKYDHNLANLNCPEFISFYYDQFMKPSEKMAPYRDFLARWSEILGFDLVQMPALAYMFYDVYTAQLGLGVPLDEATAQKFPEIEAAAGAAIDVVFGDENYIALQAGVLLNEFLKAAAAVVGGESARRVHVYSAHDYNVYSLMAVSRISPRQGVPKYGSVFSLELRKVTATGKYVVLPVYLRSPYEQIQYLQVEGCSALLCDLEEFQQLTSRYVLEEDEWRTKCGFTHDLEVDDSSIN
Summary
Description
A non-specific tyrosine phosphatase that dephosphorylates a diverse number of substrates under acidic conditions (pH 4-6) including alkyl, aryl, and acyl orthophosphate monoesters and phosphorylated proteins. Has lipid phosphatase activity and inactivates lysophosphatidic acid in seminal plasma (By similarity).
Isoform 2: the cellular form also has ecto-5'-nucleotidase activity in dorsal root ganglion (DRG) neurons. Generates adenosine from AMP which acts as a pain suppressor.
Isoform 2: the cellular form also has ecto-5'-nucleotidase activity in dorsal root ganglion (DRG) neurons. Generates adenosine from AMP which acts as a pain suppressor.
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
Subunit
Homodimer; dimer formation is required for phosphatase activity.
Similarity
Belongs to the histidine acid phosphatase family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Lysosome
Membrane
Reference proteome
Secreted
Signal
Feature
chain Prostatic acid phosphatase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JXV1
A0A437B8H1
A0A2A4K724
A0A2H1W2M4
A0A2H1W2H8
A0A2A4K8E9
+ More
A0A2A4K851 I4DJY6 A0A194PY59 A0A0N1IF44 A0A1E1WMB7 H9JIY7 I4DR67 A0A437AY25 S4NSI3 A0A1E1W159 A0A194PS41 A0A194R7T8 A0A2A4JFS4 A0A3S2LGG3 A0A212EPG3 H9J9I3 A0A2H1VAX3 A0A2A4K986 A0A212F630 A0A0N0PBK2 A0A194PY67 A0A2H1WGF6 K7IQ61 A0A232EEI8 A0A250YH28 I3N6C9 G1SYZ1 A0A2K6FQ70 A0A0L7LP95 A0A1Y1KWW5 A0A1Y1KWT0 F1Q4D3 A0A1U7V1N2 A0A2U3X1Y4 G3WUE8 G3WUE9 Q8CE08 A0A452EWS2 A0A2K5RDL6 Q8CE08-2 A0A3Q7UDM5 A0A1U7RXA5 U3KDB9 A0A151P351 A0A1U8CY71 A0A3Q0CY15 A0A2K6SJD8 K9IJ78 A0A1U7QX17 U3BAN4 U3BVD3 A0A2K5DU51 Q3I1Z6 A6H730 W5PH63 A0A2K5KUD1 K9IXP4 Q3I1Y9 A0A0D9RDS3 F6Y5Y5 A0A1D5QQZ0 A0A093I7X9 G3STE9 K7IQ63 H2PBF8 A0A1S3WUQ9 A0A287AFE9 A0A2J8U3K3 D2A428 A0A0G2K4B4 A0A2K5I5Z0 F1SPF0 A0A3Q1M6R0 A0A2K6RIZ6 Q3I1Z7 A0A2R8N8D9 A0A2U3XZS1 A0A2Y9G6Q0 Q3I1Z9 Q3I1Z8 L8IIS0 G5BKQ9 A0A2Y9G9V7 A0A093ERJ0 G7NXZ7 A0A0G2JSL5 A0A3Q7Q7F7 A0A3Q7PWT8 A0A3Q2HX16 H2R438
A0A2A4K851 I4DJY6 A0A194PY59 A0A0N1IF44 A0A1E1WMB7 H9JIY7 I4DR67 A0A437AY25 S4NSI3 A0A1E1W159 A0A194PS41 A0A194R7T8 A0A2A4JFS4 A0A3S2LGG3 A0A212EPG3 H9J9I3 A0A2H1VAX3 A0A2A4K986 A0A212F630 A0A0N0PBK2 A0A194PY67 A0A2H1WGF6 K7IQ61 A0A232EEI8 A0A250YH28 I3N6C9 G1SYZ1 A0A2K6FQ70 A0A0L7LP95 A0A1Y1KWW5 A0A1Y1KWT0 F1Q4D3 A0A1U7V1N2 A0A2U3X1Y4 G3WUE8 G3WUE9 Q8CE08 A0A452EWS2 A0A2K5RDL6 Q8CE08-2 A0A3Q7UDM5 A0A1U7RXA5 U3KDB9 A0A151P351 A0A1U8CY71 A0A3Q0CY15 A0A2K6SJD8 K9IJ78 A0A1U7QX17 U3BAN4 U3BVD3 A0A2K5DU51 Q3I1Z6 A6H730 W5PH63 A0A2K5KUD1 K9IXP4 Q3I1Y9 A0A0D9RDS3 F6Y5Y5 A0A1D5QQZ0 A0A093I7X9 G3STE9 K7IQ63 H2PBF8 A0A1S3WUQ9 A0A287AFE9 A0A2J8U3K3 D2A428 A0A0G2K4B4 A0A2K5I5Z0 F1SPF0 A0A3Q1M6R0 A0A2K6RIZ6 Q3I1Z7 A0A2R8N8D9 A0A2U3XZS1 A0A2Y9G6Q0 Q3I1Z9 Q3I1Z8 L8IIS0 G5BKQ9 A0A2Y9G9V7 A0A093ERJ0 G7NXZ7 A0A0G2JSL5 A0A3Q7Q7F7 A0A3Q7PWT8 A0A3Q2HX16 H2R438
EC Number
3.1.3.2
Pubmed
19121390
22651552
26354079
23622113
22118469
20075255
+ More
28648823 28087693 21993624 26227816 28004739 16341006 21709235 16141072 19468303 15489334 17638863 18940592 20084276 22293439 25243066 16170411 20809919 18464734 17431167 18362917 19820115 15057822 19393038 25362486 22751099 21993625 22002653 19892987 16136131
28648823 28087693 21993624 26227816 28004739 16341006 21709235 16141072 19468303 15489334 17638863 18940592 20084276 22293439 25243066 16170411 20809919 18464734 17431167 18362917 19820115 15057822 19393038 25362486 22751099 21993625 22002653 19892987 16136131
EMBL
BABH01040368
RSAL01000117
RVE46811.1
NWSH01000059
PCG80075.1
ODYU01005909
+ More
SOQ47287.1 SOQ47289.1 PCG80073.1 PCG80074.1 AK401604 BAM18226.1 KQ459585 KPI98276.1 KQ460778 KPJ12419.1 GDQN01003058 JAT87996.1 BABH01022999 BABH01023000 AK404965 BAM20407.1 RSAL01000278 RVE43078.1 GAIX01014082 JAA78478.1 GDQN01010326 JAT80728.1 KQ459601 KPI93945.1 KQ460855 KPJ11936.1 NWSH01001685 PCG70424.1 RSAL01000014 RVE53289.1 AGBW02013469 OWR43369.1 BABH01004863 ODYU01001406 SOQ37542.1 NWSH01000024 PCG80629.1 AGBW02010086 OWR49186.1 KQ460889 KPJ11041.1 KPI98277.1 ODYU01008500 SOQ52158.1 NNAY01005308 OXU16770.1 GFFW01001898 JAV42890.1 AGTP01046392 AGTP01046393 AGTP01046394 AGTP01046395 AGTP01046396 AGTP01046397 AGTP01046398 AGTP01046399 AGTP01046400 AGTP01046401 JTDY01000422 KOB77267.1 GEZM01071381 GEZM01071374 JAV65852.1 GEZM01071378 GEZM01071370 JAV65859.1 AAEX03013595 AEFK01201456 AEFK01201457 AEFK01201458 AEFK01201459 AF210243 AK029273 AK076383 CT030733 BC139826 LWLT01000001 AGTO01001245 AKHW03001210 KYO43349.1 GABZ01006540 JAA46985.1 GAMT01002023 JAB09838.1 GAMT01002022 JAB09839.1 DQ150472 AAZ82242.1 BC146093 AMGL01011780 AMGL01011781 AMGL01011782 GABZ01006299 JAA47226.1 DQ150475 AAZ82249.1 AQIB01076356 AQIB01076357 AQIB01076358 JSUE03024841 KL206987 KFV87975.1 ABGA01305496 ABGA01305497 ABGA01305498 ABGA01305499 ABGA01305500 ABGA01305501 ABGA01305502 NDHI03003471 PNJ39852.1 AEMK02000088 PNJ39853.1 KQ971348 EFA05605.1 AABR07071383 DQ150471 AAZ82241.1 DQ150469 AAZ82239.1 DQ150470 AAZ82240.1 JH881123 ELR56450.1 JH170773 GEBF01004118 EHB09870.1 JAN99514.1 KK369956 KFV43052.1 AQIA01037926 CM001277 EHH51672.1 AACZ04048175
SOQ47287.1 SOQ47289.1 PCG80073.1 PCG80074.1 AK401604 BAM18226.1 KQ459585 KPI98276.1 KQ460778 KPJ12419.1 GDQN01003058 JAT87996.1 BABH01022999 BABH01023000 AK404965 BAM20407.1 RSAL01000278 RVE43078.1 GAIX01014082 JAA78478.1 GDQN01010326 JAT80728.1 KQ459601 KPI93945.1 KQ460855 KPJ11936.1 NWSH01001685 PCG70424.1 RSAL01000014 RVE53289.1 AGBW02013469 OWR43369.1 BABH01004863 ODYU01001406 SOQ37542.1 NWSH01000024 PCG80629.1 AGBW02010086 OWR49186.1 KQ460889 KPJ11041.1 KPI98277.1 ODYU01008500 SOQ52158.1 NNAY01005308 OXU16770.1 GFFW01001898 JAV42890.1 AGTP01046392 AGTP01046393 AGTP01046394 AGTP01046395 AGTP01046396 AGTP01046397 AGTP01046398 AGTP01046399 AGTP01046400 AGTP01046401 JTDY01000422 KOB77267.1 GEZM01071381 GEZM01071374 JAV65852.1 GEZM01071378 GEZM01071370 JAV65859.1 AAEX03013595 AEFK01201456 AEFK01201457 AEFK01201458 AEFK01201459 AF210243 AK029273 AK076383 CT030733 BC139826 LWLT01000001 AGTO01001245 AKHW03001210 KYO43349.1 GABZ01006540 JAA46985.1 GAMT01002023 JAB09838.1 GAMT01002022 JAB09839.1 DQ150472 AAZ82242.1 BC146093 AMGL01011780 AMGL01011781 AMGL01011782 GABZ01006299 JAA47226.1 DQ150475 AAZ82249.1 AQIB01076356 AQIB01076357 AQIB01076358 JSUE03024841 KL206987 KFV87975.1 ABGA01305496 ABGA01305497 ABGA01305498 ABGA01305499 ABGA01305500 ABGA01305501 ABGA01305502 NDHI03003471 PNJ39852.1 AEMK02000088 PNJ39853.1 KQ971348 EFA05605.1 AABR07071383 DQ150471 AAZ82241.1 DQ150469 AAZ82239.1 DQ150470 AAZ82240.1 JH881123 ELR56450.1 JH170773 GEBF01004118 EHB09870.1 JAN99514.1 KK369956 KFV43052.1 AQIA01037926 CM001277 EHH51672.1 AACZ04048175
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000002358 UP000215335 UP000005215 UP000001811 UP000233160 UP000037510 UP000002254 UP000189704 UP000245340 UP000007648 UP000000589 UP000291000 UP000233040 UP000286640 UP000189705 UP000016665 UP000050525 UP000189706 UP000233220 UP000233020 UP000009136 UP000002356 UP000233060 UP000029965 UP000002279 UP000006718 UP000053584 UP000007646 UP000001595 UP000079721 UP000008227 UP000007266 UP000002494 UP000233080 UP000233200 UP000008225 UP000245341 UP000248481 UP000006813 UP000009130 UP000233100 UP000286641 UP000002281 UP000002277
UP000002358 UP000215335 UP000005215 UP000001811 UP000233160 UP000037510 UP000002254 UP000189704 UP000245340 UP000007648 UP000000589 UP000291000 UP000233040 UP000286640 UP000189705 UP000016665 UP000050525 UP000189706 UP000233220 UP000233020 UP000009136 UP000002356 UP000233060 UP000029965 UP000002279 UP000006718 UP000053584 UP000007646 UP000001595 UP000079721 UP000008227 UP000007266 UP000002494 UP000233080 UP000233200 UP000008225 UP000245341 UP000248481 UP000006813 UP000009130 UP000233100 UP000286641 UP000002281 UP000002277
Interpro
Gene 3D
ProteinModelPortal
H9JXV1
A0A437B8H1
A0A2A4K724
A0A2H1W2M4
A0A2H1W2H8
A0A2A4K8E9
+ More
A0A2A4K851 I4DJY6 A0A194PY59 A0A0N1IF44 A0A1E1WMB7 H9JIY7 I4DR67 A0A437AY25 S4NSI3 A0A1E1W159 A0A194PS41 A0A194R7T8 A0A2A4JFS4 A0A3S2LGG3 A0A212EPG3 H9J9I3 A0A2H1VAX3 A0A2A4K986 A0A212F630 A0A0N0PBK2 A0A194PY67 A0A2H1WGF6 K7IQ61 A0A232EEI8 A0A250YH28 I3N6C9 G1SYZ1 A0A2K6FQ70 A0A0L7LP95 A0A1Y1KWW5 A0A1Y1KWT0 F1Q4D3 A0A1U7V1N2 A0A2U3X1Y4 G3WUE8 G3WUE9 Q8CE08 A0A452EWS2 A0A2K5RDL6 Q8CE08-2 A0A3Q7UDM5 A0A1U7RXA5 U3KDB9 A0A151P351 A0A1U8CY71 A0A3Q0CY15 A0A2K6SJD8 K9IJ78 A0A1U7QX17 U3BAN4 U3BVD3 A0A2K5DU51 Q3I1Z6 A6H730 W5PH63 A0A2K5KUD1 K9IXP4 Q3I1Y9 A0A0D9RDS3 F6Y5Y5 A0A1D5QQZ0 A0A093I7X9 G3STE9 K7IQ63 H2PBF8 A0A1S3WUQ9 A0A287AFE9 A0A2J8U3K3 D2A428 A0A0G2K4B4 A0A2K5I5Z0 F1SPF0 A0A3Q1M6R0 A0A2K6RIZ6 Q3I1Z7 A0A2R8N8D9 A0A2U3XZS1 A0A2Y9G6Q0 Q3I1Z9 Q3I1Z8 L8IIS0 G5BKQ9 A0A2Y9G9V7 A0A093ERJ0 G7NXZ7 A0A0G2JSL5 A0A3Q7Q7F7 A0A3Q7PWT8 A0A3Q2HX16 H2R438
A0A2A4K851 I4DJY6 A0A194PY59 A0A0N1IF44 A0A1E1WMB7 H9JIY7 I4DR67 A0A437AY25 S4NSI3 A0A1E1W159 A0A194PS41 A0A194R7T8 A0A2A4JFS4 A0A3S2LGG3 A0A212EPG3 H9J9I3 A0A2H1VAX3 A0A2A4K986 A0A212F630 A0A0N0PBK2 A0A194PY67 A0A2H1WGF6 K7IQ61 A0A232EEI8 A0A250YH28 I3N6C9 G1SYZ1 A0A2K6FQ70 A0A0L7LP95 A0A1Y1KWW5 A0A1Y1KWT0 F1Q4D3 A0A1U7V1N2 A0A2U3X1Y4 G3WUE8 G3WUE9 Q8CE08 A0A452EWS2 A0A2K5RDL6 Q8CE08-2 A0A3Q7UDM5 A0A1U7RXA5 U3KDB9 A0A151P351 A0A1U8CY71 A0A3Q0CY15 A0A2K6SJD8 K9IJ78 A0A1U7QX17 U3BAN4 U3BVD3 A0A2K5DU51 Q3I1Z6 A6H730 W5PH63 A0A2K5KUD1 K9IXP4 Q3I1Y9 A0A0D9RDS3 F6Y5Y5 A0A1D5QQZ0 A0A093I7X9 G3STE9 K7IQ63 H2PBF8 A0A1S3WUQ9 A0A287AFE9 A0A2J8U3K3 D2A428 A0A0G2K4B4 A0A2K5I5Z0 F1SPF0 A0A3Q1M6R0 A0A2K6RIZ6 Q3I1Z7 A0A2R8N8D9 A0A2U3XZS1 A0A2Y9G6Q0 Q3I1Z9 Q3I1Z8 L8IIS0 G5BKQ9 A0A2Y9G9V7 A0A093ERJ0 G7NXZ7 A0A0G2JSL5 A0A3Q7Q7F7 A0A3Q7PWT8 A0A3Q2HX16 H2R438
PDB
1RPT
E-value=7.05579e-29,
Score=317
Ontologies
GO
GO:0003993
GO:0016021
GO:0016311
GO:0008253
GO:0030175
GO:0006144
GO:0060168
GO:0012506
GO:0009117
GO:0006772
GO:0046085
GO:0016791
GO:0042803
GO:0005615
GO:0052642
GO:0042131
GO:0051930
GO:0005886
GO:0051289
GO:0005771
GO:0033265
GO:0045177
GO:0042802
GO:0005765
GO:0031985
GO:0030141
GO:0007165
GO:0005488
GO:0005515
GO:0005634
GO:0030145
GO:0006412
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Secreted
Cell membrane Appears to shuttle between the cell membrane and intracellular vesicles. Colocalizes with FLOT1 at cell membrane and in intracellular vesicles (PubMed:17638863). Colocalizes with LAMP2 on the lysosome membrane (By similarity). With evidence from 2 publications.
Lysosome membrane Appears to shuttle between the cell membrane and intracellular vesicles. Colocalizes with FLOT1 at cell membrane and in intracellular vesicles (PubMed:17638863). Colocalizes with LAMP2 on the lysosome membrane (By similarity). With evidence from 2 publications.
Cell membrane Appears to shuttle between the cell membrane and intracellular vesicles. Colocalizes with FLOT1 at cell membrane and in intracellular vesicles (PubMed:17638863). Colocalizes with LAMP2 on the lysosome membrane (By similarity). With evidence from 2 publications.
Lysosome membrane Appears to shuttle between the cell membrane and intracellular vesicles. Colocalizes with FLOT1 at cell membrane and in intracellular vesicles (PubMed:17638863). Colocalizes with LAMP2 on the lysosome membrane (By similarity). With evidence from 2 publications.
SignalP
Position: 1 - 19,
Likelihood: 0.992620
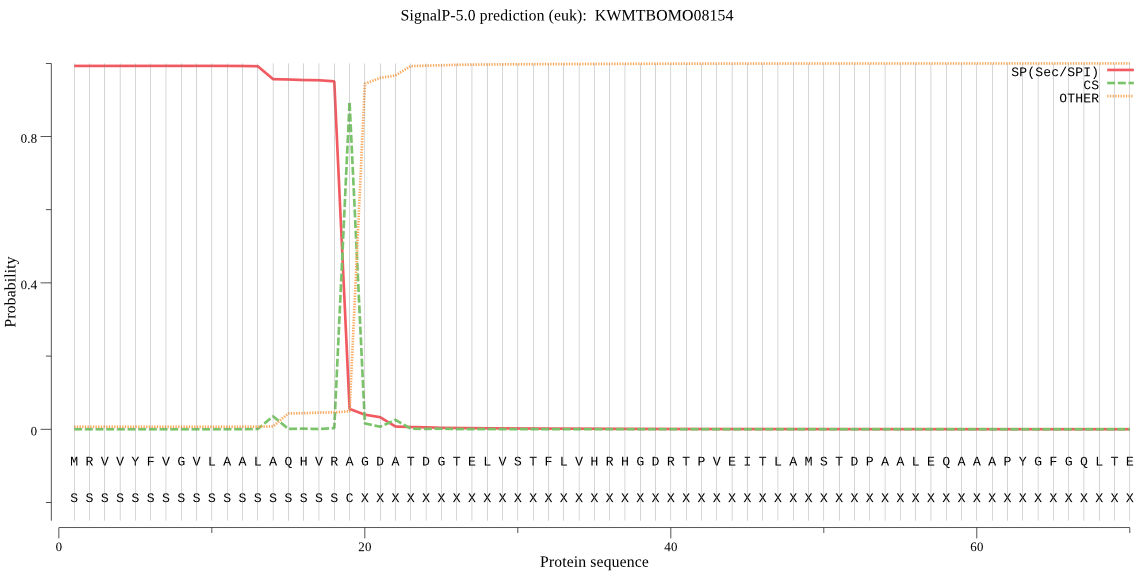
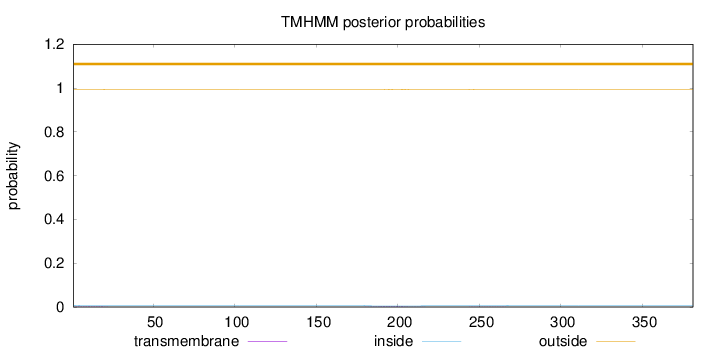
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12066
Exp number, first 60 AAs:
0.02862
Total prob of N-in:
0.00795
outside
1 - 381
Population Genetic Test Statistics
Pi
226.242586
Theta
182.135115
Tajima's D
0.662565
CLR
2.356611
CSRT
0.559822008899555
Interpretation
Uncertain
