Gene
KWMTBOMO08152
Pre Gene Modal
BGIBMGA014373
Annotation
PREDICTED:_protein_toll-like_[Bombyx_mori]
Full name
Protein toll
Location in the cell
Nuclear Reliability : 1.08 PlasmaMembrane Reliability : 1.192
Sequence
CDS
ATGGACCTCTGCCCCGGCAACTCCAACGCACACGTCGGCTCCGGCACTTCCCCGCTAAAAGCGCTGACGGAGTTGGATGAGCTGAGACTGGGTCACACGGGTGTGTCTCACGTGTGCAGCGACTGGCGGACCTCCATGAAGCAACTCAGTCTTCTCGATCTCTCCTACACAAACATCACCTCCCTGTCGTACCTGGATTTCCACTACGAGCGCTCTGGATCCAAAAAGCTGACCGTGAACTTCAACGGCACGCCCGTCAGCGAGCTCACCGTCGACGCGGGGAACTACAAGTCTGTGATCGCCGACCTGGCATCTGCATCGGTCAAGATAATATTGAGAGCAACTTTACCGTGCGACTGCTTGGACTACTGGACGGCACTGGCGTTCGGGAACCTCAGCACTCAAGCACAAGATTCCGTGGAGCTCCTGTGTAGGGAGGAGGAGCGTGGTTCACTAGCGGACGCGTTGGAACGGGAATCTCTGGAGTGTGCGGAATCGAGCCTGTGTGAGGACCAGGAAGGCTGCACCTGTTCCGTACGCCCTGACCTCTCGCATGGTGTGGTGGCGACCGCGTCATGTGAGGGCGCGGGTCTGGACGAAATGCCATCGCGTCTGAAGTTCAAAGAGGCTCCCGCGTGGCGCTTAATGCTCGCCCACAACCACATCCAGACCGTTCGGTTTCAGGATCTGCCCGACACGATATTGGAGTTGGACCTGAGAAACAACTCTCTGCATAGATTGGACGGCCCGACTGCTTCGAAGTTATCAACTGTCCCTCTGTGGATCGCCAACAATTCGCTGGACTGCACATGTCACGGGTACGACTTGGTGACGAACCTCGAACTGTGTGTGCTCGACATGGAGCAAGCGACGTGTGACGACGGGACCCCTCTAATTGAAGCACAGATCAACGATCCCACTGAATGCAGTTCGTTTGTAACGGTCACCGTCTCTTTGATCAGCGCGCTGGCTTTGTTGGCAGTGATGGTCGCAGTTACAGCCGGCTGCGGGATGCGACCCGAGATGCGGCTGAGAGTTAAGGTGCTGCTGCTGCGACTGGGCTGGCTGCCACGGCGGCCGGAGCCGGACGACGGGCGTCGGTTCGATGCGTTCGTATCGTACGCGCACGAGGACGAGGCGGTGGTGGAAGAGCTCGTGAAGCGGCTTGAGGTGGGGCACGGCTACCGGCTGTGCCTGCACTACCGCGACTGGCCGCCGGGCGAGTGGATCCACGTGCAGATCGCGGCCTCGGTGCAAGCGGCGCGCCGCACCCTCATCGTAGTCTCGCGGCACTTCCTGCGCTCCAAGTGGGCGCGCCAAGAGTTCCGCCAGGCACACGCGGCCGCGCTGCGTGACGTCACACCGCGCCTCGTGCTGCTGTTCCTCGAGCCGCCGCACCGCTTGCCTCTCGACGCCGAGCTGCGCTCCTACATTAGGATCAACACCTACCTGCTCTGGACCGACCCCTGGTTCTGGCACAAACTAAAGCTAGCCCTGCCGCCCCCGCGCCTGCTCCCCACCCCGTTCGAGGATGTCTCCGTTAATAAGGCGGAGAGCTCCCCTCTACAAGTAGCGCCACGCCCGCAGAGTGCGTGA
Protein
MDLCPGNSNAHVGSGTSPLKALTELDELRLGHTGVSHVCSDWRTSMKQLSLLDLSYTNITSLSYLDFHYERSGSKKLTVNFNGTPVSELTVDAGNYKSVIADLASASVKIILRATLPCDCLDYWTALAFGNLSTQAQDSVELLCREEERGSLADALERESLECAESSLCEDQEGCTCSVRPDLSHGVVATASCEGAGLDEMPSRLKFKEAPAWRLMLAHNHIQTVRFQDLPDTILELDLRNNSLHRLDGPTASKLSTVPLWIANNSLDCTCHGYDLVTNLELCVLDMEQATCDDGTPLIEAQINDPTECSSFVTVTVSLISALALLAVMVAVTAGCGMRPEMRLRVKVLLLRLGWLPRRPEPDDGRRFDAFVSYAHEDEAVVEELVKRLEVGHGYRLCLHYRDWPPGEWIHVQIAASVQAARRTLIVVSRHFLRSKWARQEFRQAHAAALRDVTPRLVLLFLEPPHRLPLDAELRSYIRINTYLLWTDPWFWHKLKLALPPPRLLPTPFEDVSVNKAESSPLQVAPRPQSA
Summary
Description
Receptor for the cleaved activated form of spz, spaetzle C-106 (PubMed:12872120). Binding to spaetzle C-106 activates the Toll signaling pathway and induces expression of the antifungal peptide drosomycin (PubMed:12872120, PubMed:8808632, PubMed:10973475). Component of the extracellular signaling pathway that establishes dorsal-ventral polarity in the embryo (PubMed:3931919). Promotes heterophilic cellular adhesion (PubMed:2124970). Involved in synaptic targeting of motoneurons RP5 and V to muscle 12 (M12); functions as a repulsive cue inhibiting motoneuron synapse formation on muscle 13 (M13) to guide RP5 and V to the neighboring M12, where its expression is repressed by tey (PubMed:20504957). May also function in embryonic neuronal survival and the synaptic targeting of SNa motoneurons (PubMed:19018662).
Subunit
In the absence of ligand, forms a low-affinity disulfide-linked homodimer (PubMed:24733933). In the presence of ligand, crystal structures show one Tl molecule bound to a spaetzle C-106 homodimer (PubMed:24282309, PubMed:24733933). However, the active complex probably consists of two Tl molecules bound to a spaetzle C-106 homodimer (PubMed:24282309, PubMed:24733933). This is supported by in vitro experiments which also show binding of the spaetzle C-106 dimer to 2 Tl receptors (PubMed:12872120). Ligand binding induces conformational changes in the extracellular domain of Tl (PubMed:24282309). This may enable a secondary homodimerization interface at the C-terminus of the Tl extracellular domain (PubMed:24282309).
Similarity
Belongs to the Toll-like receptor family.
Keywords
3D-structure
Cell adhesion
Cell membrane
Complete proteome
Cytoplasm
Developmental protein
Disulfide bond
Glycoprotein
Immunity
Innate immunity
Leucine-rich repeat
Membrane
Polymorphism
Receptor
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Protein toll
Uniprot
H9JXV5
H9JXV2
A0A2H1W8Q3
A4GVU0
A0A0L7LPX8
A0A0M8ZY10
+ More
A0A0P4VMK0 T1HYZ2 A0A0L7QLM2 A0A224X644 A0A195F0C9 A0A026W7S0 A0A346RAF5 A0A151JAL5 E2BBY4 F4W4T1 A0A158P3M6 A0A310S7W9 A0A154NYN7 A1C1P2 D2CGM6 A0A2J7QHL9 A0A067R3H7 E5D2L0 A0A088AG15 B2WSC9 A0A2J7QHL1 D7EZ50 B4K8Y1 A0A232FI58 A0A2A3EQD4 K7IRA7 Q8MQU7 B5BRC1 A0A0A9XC66 A0A146L775 W8PVP4 A0A410SF83 A0A0D3QDL8 A0A182GPG4 A0A0D3QDD7 B3P637 V9I193 A0A2C9H6H6 B4LWB8 A0A182IDR8 B4PSC7 Q6V666 Q24622 Q6V669 B4QWS2 A0A1S4FH10 Q6V672 Q56RA0 A0A1B0CQQ9 Q6V670 A0A3B0K7X3 A0A2C9GWS9 A0A0M5J9C0 A0A182UFK6 A0A0D3QD03 A0A0D3QDC7 Q8WRE5 Q6V495 A0A0D3QDL3 A0A0D3QCR2 A0A0D3QCQ2 Q6V673 Q6V667 Q56R98 A0A0B4KHY4 T2GGK5 P08953 Q29B96 W8PWC1 A0A0D3QCQ7 Q8MQU9 A0A182Y4Y2 A0A182PVM9 K7ITQ2 A0A1L8E2X4 Q17F73 A0A1I8PSW5 A0A1S4F4W7 A0A182M8X8 Q6V671 J9JPZ4 A0A182XNF0 A0A182KC65
A0A0P4VMK0 T1HYZ2 A0A0L7QLM2 A0A224X644 A0A195F0C9 A0A026W7S0 A0A346RAF5 A0A151JAL5 E2BBY4 F4W4T1 A0A158P3M6 A0A310S7W9 A0A154NYN7 A1C1P2 D2CGM6 A0A2J7QHL9 A0A067R3H7 E5D2L0 A0A088AG15 B2WSC9 A0A2J7QHL1 D7EZ50 B4K8Y1 A0A232FI58 A0A2A3EQD4 K7IRA7 Q8MQU7 B5BRC1 A0A0A9XC66 A0A146L775 W8PVP4 A0A410SF83 A0A0D3QDL8 A0A182GPG4 A0A0D3QDD7 B3P637 V9I193 A0A2C9H6H6 B4LWB8 A0A182IDR8 B4PSC7 Q6V666 Q24622 Q6V669 B4QWS2 A0A1S4FH10 Q6V672 Q56RA0 A0A1B0CQQ9 Q6V670 A0A3B0K7X3 A0A2C9GWS9 A0A0M5J9C0 A0A182UFK6 A0A0D3QD03 A0A0D3QDC7 Q8WRE5 Q6V495 A0A0D3QDL3 A0A0D3QCR2 A0A0D3QCQ2 Q6V673 Q6V667 Q56R98 A0A0B4KHY4 T2GGK5 P08953 Q29B96 W8PWC1 A0A0D3QCQ7 Q8MQU9 A0A182Y4Y2 A0A182PVM9 K7ITQ2 A0A1L8E2X4 Q17F73 A0A1I8PSW5 A0A1S4F4W7 A0A182M8X8 Q6V671 J9JPZ4 A0A182XNF0 A0A182KC65
Pubmed
19121390
17606296
26227816
27129103
24508170
30249741
+ More
29910977 20798317 21719571 21347285 17056116 24845553 18321728 17994087 28648823 20075255 12542637 25401762 26823975 25552603 26483478 24398262 18057021 17550304 12930753 8112609 17510324 15545653 16547107 12213252 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2449285 12537569 3931919 2124970 1879347 8808632 10973475 12617819 12872120 15373972 19018662 22464169 20504957 23596340 24282309 23245851 24733933 15632085 25244985
29910977 20798317 21719571 21347285 17056116 24845553 18321728 17994087 28648823 20075255 12542637 25401762 26823975 25552603 26483478 24398262 18057021 17550304 12930753 8112609 17510324 15545653 16547107 12213252 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2449285 12537569 3931919 2124970 1879347 8808632 10973475 12617819 12872120 15373972 19018662 22464169 20504957 23596340 24282309 23245851 24733933 15632085 25244985
EMBL
BABH01040363
BABH01040364
BABH01040360
BABH01040361
ODYU01007053
SOQ49485.1
+ More
EF442782 ABO21763.1 JTDY01000372 KOB77487.1 KQ435801 KOX73247.1 GDKW01003048 JAI53547.1 ACPB03009442 KQ414914 KOC59528.1 GFTR01008481 JAW07945.1 KQ981880 KYN33933.1 KK107356 QOIP01000010 EZA52048.1 RLU17569.1 MH243021 AXS59155.1 KQ979262 KYN22067.1 GL447175 EFN86808.1 GL887553 EGI70800.1 ADTU01008265 KQ775195 OAD52269.1 KQ434773 KZC03990.1 DQ923424 ABK58729.1 EF117252 ABO38434.1 NEVH01013973 PNF28084.1 KK852931 KDR13629.1 GU014556 ADK55066.1 EF407561 ABQ59330.1 PNF28085.1 EU433386 ACC68670.1 CH933806 EDW16578.1 NNAY01000160 OXU30431.1 KZ288197 PBC33714.1 AY124197 AAM97776.1 AB385869 BAG68890.1 GBHO01026374 GBHO01026373 GBHO01026372 GBHO01026370 JAG17230.1 JAG17231.1 JAG17232.1 JAG17234.1 GDHC01015553 GDHC01008551 JAQ03076.1 JAQ10078.1 KJ188411 AHL39101.1 MH178390 QAT92918.1 KP274553 AJC98699.1 JXUM01078342 JXUM01078343 KP274549 AJC98695.1 CH954182 EDV53507.1 JF895474 AEI25533.1 CH940650 EDW67652.2 KRF83383.1 APCN01004779 APCN01004780 CM000160 EDW98589.2 AY349924 AAQ64875.1 L25390 AAA28942.1 AY349921 AAQ64872.1 CM000364 EDX14589.1 AY349918 AAQ64869.1 AY754607 AAX13182.1 AJWK01023878 AY349920 AAQ64871.1 OUUW01000005 SPP81101.1 CP012526 ALC46724.1 KP274543 KP274546 AJC98689.1 AJC98692.1 KP274533 KP274534 KP274535 KP274536 KP274537 KP274538 KP274539 KP274540 KP274541 KP274544 KP274551 AJC98679.1 AJC98680.1 AJC98681.1 AJC98682.1 AJC98683.1 AJC98684.1 AJC98685.1 AJC98686.1 AJC98687.1 AJC98690.1 AJC98697.1 AF444780 AAAB01008811 AAL37901.1 EAA45376.1 AY352264 AAQ65064.1 KP274545 KP274548 KP274550 AJC98691.1 AJC98694.1 AJC98696.1 KP274552 AJC98698.1 KP274542 AJC98688.1 AY349917 AAQ64868.1 AY349922 AY349923 AAQ64873.1 AAQ64874.1 AY754609 AAX13184.1 AE014297 AGB96375.1 BT150335 AGW25617.1 M19969 BT031153 AY349649 AY349650 AY349651 AY349652 AY349653 AY349654 AY349655 AY121616 CM000070 EAL27102.2 KJ188410 AHL39100.1 KP274547 AJC98693.1 AY124195 AAM97774.1 GFDF01000994 JAV13090.1 CH477273 EAT45212.2 AXCM01000253 AY349919 AAQ64870.1 ABLF02029217
EF442782 ABO21763.1 JTDY01000372 KOB77487.1 KQ435801 KOX73247.1 GDKW01003048 JAI53547.1 ACPB03009442 KQ414914 KOC59528.1 GFTR01008481 JAW07945.1 KQ981880 KYN33933.1 KK107356 QOIP01000010 EZA52048.1 RLU17569.1 MH243021 AXS59155.1 KQ979262 KYN22067.1 GL447175 EFN86808.1 GL887553 EGI70800.1 ADTU01008265 KQ775195 OAD52269.1 KQ434773 KZC03990.1 DQ923424 ABK58729.1 EF117252 ABO38434.1 NEVH01013973 PNF28084.1 KK852931 KDR13629.1 GU014556 ADK55066.1 EF407561 ABQ59330.1 PNF28085.1 EU433386 ACC68670.1 CH933806 EDW16578.1 NNAY01000160 OXU30431.1 KZ288197 PBC33714.1 AY124197 AAM97776.1 AB385869 BAG68890.1 GBHO01026374 GBHO01026373 GBHO01026372 GBHO01026370 JAG17230.1 JAG17231.1 JAG17232.1 JAG17234.1 GDHC01015553 GDHC01008551 JAQ03076.1 JAQ10078.1 KJ188411 AHL39101.1 MH178390 QAT92918.1 KP274553 AJC98699.1 JXUM01078342 JXUM01078343 KP274549 AJC98695.1 CH954182 EDV53507.1 JF895474 AEI25533.1 CH940650 EDW67652.2 KRF83383.1 APCN01004779 APCN01004780 CM000160 EDW98589.2 AY349924 AAQ64875.1 L25390 AAA28942.1 AY349921 AAQ64872.1 CM000364 EDX14589.1 AY349918 AAQ64869.1 AY754607 AAX13182.1 AJWK01023878 AY349920 AAQ64871.1 OUUW01000005 SPP81101.1 CP012526 ALC46724.1 KP274543 KP274546 AJC98689.1 AJC98692.1 KP274533 KP274534 KP274535 KP274536 KP274537 KP274538 KP274539 KP274540 KP274541 KP274544 KP274551 AJC98679.1 AJC98680.1 AJC98681.1 AJC98682.1 AJC98683.1 AJC98684.1 AJC98685.1 AJC98686.1 AJC98687.1 AJC98690.1 AJC98697.1 AF444780 AAAB01008811 AAL37901.1 EAA45376.1 AY352264 AAQ65064.1 KP274545 KP274548 KP274550 AJC98691.1 AJC98694.1 AJC98696.1 KP274552 AJC98698.1 KP274542 AJC98688.1 AY349917 AAQ64868.1 AY349922 AY349923 AAQ64873.1 AAQ64874.1 AY754609 AAX13184.1 AE014297 AGB96375.1 BT150335 AGW25617.1 M19969 BT031153 AY349649 AY349650 AY349651 AY349652 AY349653 AY349654 AY349655 AY121616 CM000070 EAL27102.2 KJ188410 AHL39100.1 KP274547 AJC98693.1 AY124195 AAM97774.1 GFDF01000994 JAV13090.1 CH477273 EAT45212.2 AXCM01000253 AY349919 AAQ64870.1 ABLF02029217
Proteomes
UP000005204
UP000037510
UP000053105
UP000015103
UP000053825
UP000078541
+ More
UP000053097 UP000279307 UP000078492 UP000008237 UP000007755 UP000005205 UP000076502 UP000235965 UP000027135 UP000005203 UP000009192 UP000215335 UP000242457 UP000002358 UP000075903 UP000069940 UP000008711 UP000075920 UP000008792 UP000075840 UP000002282 UP000000304 UP000092461 UP000268350 UP000075900 UP000092553 UP000075902 UP000007062 UP000000803 UP000001819 UP000076408 UP000075885 UP000008820 UP000095300 UP000075883 UP000007819 UP000076407 UP000075881
UP000053097 UP000279307 UP000078492 UP000008237 UP000007755 UP000005205 UP000076502 UP000235965 UP000027135 UP000005203 UP000009192 UP000215335 UP000242457 UP000002358 UP000075903 UP000069940 UP000008711 UP000075920 UP000008792 UP000075840 UP000002282 UP000000304 UP000092461 UP000268350 UP000075900 UP000092553 UP000075902 UP000007062 UP000000803 UP000001819 UP000076408 UP000075885 UP000008820 UP000095300 UP000075883 UP000007819 UP000076407 UP000075881
Pfam
Interpro
SUPFAM
SSF52200
SSF52200
Gene 3D
ProteinModelPortal
H9JXV5
H9JXV2
A0A2H1W8Q3
A4GVU0
A0A0L7LPX8
A0A0M8ZY10
+ More
A0A0P4VMK0 T1HYZ2 A0A0L7QLM2 A0A224X644 A0A195F0C9 A0A026W7S0 A0A346RAF5 A0A151JAL5 E2BBY4 F4W4T1 A0A158P3M6 A0A310S7W9 A0A154NYN7 A1C1P2 D2CGM6 A0A2J7QHL9 A0A067R3H7 E5D2L0 A0A088AG15 B2WSC9 A0A2J7QHL1 D7EZ50 B4K8Y1 A0A232FI58 A0A2A3EQD4 K7IRA7 Q8MQU7 B5BRC1 A0A0A9XC66 A0A146L775 W8PVP4 A0A410SF83 A0A0D3QDL8 A0A182GPG4 A0A0D3QDD7 B3P637 V9I193 A0A2C9H6H6 B4LWB8 A0A182IDR8 B4PSC7 Q6V666 Q24622 Q6V669 B4QWS2 A0A1S4FH10 Q6V672 Q56RA0 A0A1B0CQQ9 Q6V670 A0A3B0K7X3 A0A2C9GWS9 A0A0M5J9C0 A0A182UFK6 A0A0D3QD03 A0A0D3QDC7 Q8WRE5 Q6V495 A0A0D3QDL3 A0A0D3QCR2 A0A0D3QCQ2 Q6V673 Q6V667 Q56R98 A0A0B4KHY4 T2GGK5 P08953 Q29B96 W8PWC1 A0A0D3QCQ7 Q8MQU9 A0A182Y4Y2 A0A182PVM9 K7ITQ2 A0A1L8E2X4 Q17F73 A0A1I8PSW5 A0A1S4F4W7 A0A182M8X8 Q6V671 J9JPZ4 A0A182XNF0 A0A182KC65
A0A0P4VMK0 T1HYZ2 A0A0L7QLM2 A0A224X644 A0A195F0C9 A0A026W7S0 A0A346RAF5 A0A151JAL5 E2BBY4 F4W4T1 A0A158P3M6 A0A310S7W9 A0A154NYN7 A1C1P2 D2CGM6 A0A2J7QHL9 A0A067R3H7 E5D2L0 A0A088AG15 B2WSC9 A0A2J7QHL1 D7EZ50 B4K8Y1 A0A232FI58 A0A2A3EQD4 K7IRA7 Q8MQU7 B5BRC1 A0A0A9XC66 A0A146L775 W8PVP4 A0A410SF83 A0A0D3QDL8 A0A182GPG4 A0A0D3QDD7 B3P637 V9I193 A0A2C9H6H6 B4LWB8 A0A182IDR8 B4PSC7 Q6V666 Q24622 Q6V669 B4QWS2 A0A1S4FH10 Q6V672 Q56RA0 A0A1B0CQQ9 Q6V670 A0A3B0K7X3 A0A2C9GWS9 A0A0M5J9C0 A0A182UFK6 A0A0D3QD03 A0A0D3QDC7 Q8WRE5 Q6V495 A0A0D3QDL3 A0A0D3QCR2 A0A0D3QCQ2 Q6V673 Q6V667 Q56R98 A0A0B4KHY4 T2GGK5 P08953 Q29B96 W8PWC1 A0A0D3QCQ7 Q8MQU9 A0A182Y4Y2 A0A182PVM9 K7ITQ2 A0A1L8E2X4 Q17F73 A0A1I8PSW5 A0A1S4F4W7 A0A182M8X8 Q6V671 J9JPZ4 A0A182XNF0 A0A182KC65
PDB
1O77
E-value=2.42274e-14,
Score=193
Ontologies
GO
GO:0007165
GO:0016021
GO:0045087
GO:0032991
GO:0004896
GO:0004888
GO:0035208
GO:0009897
GO:1902875
GO:0005769
GO:0007526
GO:0040015
GO:0007155
GO:0032154
GO:0007507
GO:0008063
GO:0009950
GO:0046627
GO:0045944
GO:0016201
GO:0002229
GO:0005887
GO:0019955
GO:0061760
GO:0030308
GO:0042802
GO:0009611
GO:0007416
GO:0070976
GO:0009986
GO:0002225
GO:0002804
GO:0050830
GO:0005886
GO:0005515
GO:0005488
GO:0016791
GO:0005634
GO:0030145
GO:0006412
GO:0016876
GO:0043039
Topology
Subcellular location
Cell membrane
In the fat body, detected in puntate structures along the cell periphery. Becomes evenly distributed in the cytoplasm 1 hour after bacterial infection. With evidence from 11 publications.
Cytoplasm In the fat body, detected in puntate structures along the cell periphery. Becomes evenly distributed in the cytoplasm 1 hour after bacterial infection. With evidence from 11 publications.
Cytoplasm In the fat body, detected in puntate structures along the cell periphery. Becomes evenly distributed in the cytoplasm 1 hour after bacterial infection. With evidence from 11 publications.
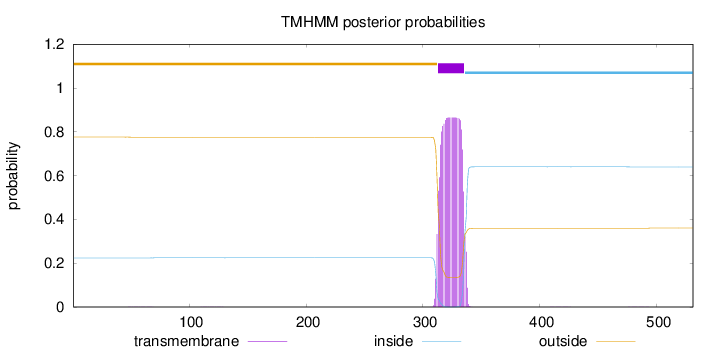
Length:
531
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.68036
Exp number, first 60 AAs:
0.00964
Total prob of N-in:
0.22389
outside
1 - 312
TMhelix
313 - 335
inside
336 - 531
Population Genetic Test Statistics
Pi
178.993261
Theta
171.854406
Tajima's D
1.167203
CLR
0.366363
CSRT
0.709414529273536
Interpretation
Uncertain
