Gene
KWMTBOMO08148
Pre Gene Modal
BGIBMGA001083
Annotation
PREDICTED:_palmitoyltransferase_ZDHHC17-like_[Papilio_machaon]
Full name
Palmitoyltransferase
+ More
Palmitoyltransferase Hip14
Palmitoyltransferase Hip14
Alternative Name
Huntingtin-interacting protein 14 homolog
Location in the cell
PlasmaMembrane Reliability : 4.023
Sequence
CDS
ATGATGTACGACAGTACTTGTGGTGCAGCGGCGACTGGCCAGTGTGGCCCTGCGCAGGCTGAACGAGAACCTCCAGAACCGCGGGAACCTCCACCACCACCAACTGAACGAGATTACAGCGGCTACGATATCGTGAAAGCAACGCAGTACGGAGCATTTGTGCGCGTTAAAGAACTCGTCGAAGCTGGCTGGGACGTGAACCGACCCGACGGCGAAACGGTGACCTTGCTGCACTGGGCTGCAATCAACAACAGACGCGACATCATCCAGTACTTGCTGTCGAAGGGTGCGAATGTCGATGCAGTTGGGGGAGAGTTACAGTCGACACCGCTACACTGGGCGACGCGGCAAGGGCACCTGGACGCGACTGTGCTGCTGGTCCGCGCGGGAGCCGACCCTTCGCTGCGTGACGCCGAGGGCTGCGCGTGCCTACACCTGGCAGCACAATTCGGTCACACTGCTGTGGTGGCGTACCTAGTGGCACGAGGTGTGACTGTGGATGAACCTGATGCAGGTGGCATGACGGCGCTCATGTGGGCAAGCTGGAAGGTTGCAGCTGTGGACCCAACACGTTTACTCCTCACTTTAGGTGCATCAACACAACCAGCTGATCGTGGCCATGGTAACACAGCCCTGCACTGGGCTATTCTGGCACGTAACACTACGGCCATATCCACTCTGATACTTTATGGTGATGCTAATTTGGATGTACCCAACCTGCGAGGAGTGACTCCATTGCAGATGTTACAGGCGAGTGCTGATTCCTTATGGGTTGGCTCTAAAGTTTCAGACAAAATAAAGGAACGTACAATGGAGTCAGAAAGATTATCAATATTTAGAAGATTGACATATGACAAGAAATTCAGATGGTGGTGTGTGGTAGTAATGCCATTCATGGCATTCTACTTGACTGGCCTCATCCTGGAGACGGACTTGAACTATCCTCTAAAAACCTTAATCTTGATTGGTTTCTACTCACTTTTACATTTTACTAGTAACTTGTTATTTGATGATGAACTCAAAAACATATTTCCCCTGAGTATATATCTCGCAACCAAGGTGTGGTTCTACATAACGTGGGTGGTGTTTATCCTTCCGGTGGTGGGGTGGAGCTGGTCTCTGGGGTTCGCAGTTTGCTCGGGGTTGCTGTGGGTGACATTCCTGCGCTCGTGGCGTTCCGATCCTGGCGTCATTACAGCTTCCAAAGCTGACAAGTTCAGGACGATCATCGAGCTGTCGGAGGCGGCGGGCGGCGGGTTCGAGTCGTCGCGGTTCTGCTCGGCGTGCCTGGTGCGGCGCCCGCTGCGGTCCAAGCACTGCGCCGTGTGCAACCGCTGCGTCGCCAAGTTCGACCACCACTGTCCCTGGGTCGGCAACTGCATCGGCGCCAACAACCACCGCCACTTCATCGGCTTCCTGTGCAGCCTCATCCTCATGTGCTGCTGGATGCTGTGGGGCGGCGCGCGCTACCTGCGCGACCAGTGCCCCGCGCCCGCCGAGCCGCTCGCCTGGGTCTCCTGCAACGGCTGGATCTCGTGGGTGATGCTCAATGCCGCGTTCCACCTGTTCTGGGTGACGGTGCTCACCGGCTTCCAGATGTACCTGGTGGTGTGCCTCGGCATGACCACCAACGAGCAGCTGAACCGCGGCCGCTACAAGCACTTCCAGGCGCGCGGCGGCCGCTCGCCCTTCAGCCGCGGCCCGCTCGCCAACTGCTTGGAGCTGCTCGGCTGCCGGTCGCCCCGGGACCACGCCGACCTGGACCACGGCGTGTGA
Protein
MMYDSTCGAAATGQCGPAQAEREPPEPREPPPPPTERDYSGYDIVKATQYGAFVRVKELVEAGWDVNRPDGETVTLLHWAAINNRRDIIQYLLSKGANVDAVGGELQSTPLHWATRQGHLDATVLLVRAGADPSLRDAEGCACLHLAAQFGHTAVVAYLVARGVTVDEPDAGGMTALMWASWKVAAVDPTRLLLTLGASTQPADRGHGNTALHWAILARNTTAISTLILYGDANLDVPNLRGVTPLQMLQASADSLWVGSKVSDKIKERTMESERLSIFRRLTYDKKFRWWCVVVMPFMAFYLTGLILETDLNYPLKTLILIGFYSLLHFTSNLLFDDELKNIFPLSIYLATKVWFYITWVVFILPVVGWSWSLGFAVCSGLLWVTFLRSWRSDPGVITASKADKFRTIIELSEAAGGGFESSRFCSACLVRRPLRSKHCAVCNRCVAKFDHHCPWVGNCIGANNHRHFIGFLCSLILMCCWMLWGGARYLRDQCPAPAEPLAWVSCNGWISWVMLNAAFHLFWVTVLTGFQMYLVVCLGMTTNEQLNRGRYKHFQARGGRSPFSRGPLANCLELLGCRSPRDHADLDHGV
Summary
Description
Probable palmitoyltransferase which is required for photoreceptor synaptic transmission and for the correct expression and localization of palmitoylated protein Csp and synaptosomal-associated protein Snap25 (PubMed:18158335, PubMed:18032660). Probably palmitoylates Csp (PubMed:18158335). Probably also palmitoylates the dorsal-ventral patterning protein Sog and promotes its secretion and activity and the stabilization of the membrane-bound form (PubMed:20599894). Required for synaptic vesicle exocytosis (PubMed:18158335).
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Subunit
Interacts with dorsal-ventral patterning protein Sog.
Similarity
Belongs to the DHHC palmitoyltransferase family.
Belongs to the DHHC palmitoyltransferase family. AKR/ZDHHC17 subfamily.
Belongs to the DHHC palmitoyltransferase family. AKR/ZDHHC17 subfamily.
Keywords
Acyltransferase
ANK repeat
Cell junction
Cell membrane
Complete proteome
Golgi apparatus
Lipoprotein
Membrane
Palmitate
Reference proteome
Repeat
Synapse
Transferase
Transmembrane
Transmembrane helix
Feature
chain Palmitoyltransferase Hip14
Uniprot
H9IV03
A0A2H1VR64
A0A2A4JGZ5
A0A194REK0
A0A2W1B5J2
A0A194QCD4
+ More
A0A182FKP1 A0A2M4APL5 W5JDI0 A0A182NPQ1 A0A182QDB5 A0A182P6X8 A0A182RIG0 A0A182YL43 A0A182SZY8 A0A182WEC7 A0A182JZP8 A0A182M6N3 A0A084VIP0 A0A1Q3FVR8 A0A1Q3FW47 A0A182INR1 A0A182G5W6 A0A2Y9D3G6 A0A182V4Y4 A0A182L4K3 Q7PZ51 A0A1I8P9M2 A0A336K7Y1 B4N727 M9PFE4 Q9VUW9 A0A1A9XT26 A0A1B0B966 Q29D93 B4H7T3 W8BBQ0 B4ITV5 B3M9T9 B3NDQ1 A0A034VE00 A0A1A9WGH9 B4QLS4 A0A1B0FAV5 B4KYH8 A0A0K8UBM9 A0A1W4VAE9 A0A0K8U1C2 A0A0K8UW18 B4J3N8 A0A0M3QWV8 A0A1B0AIE9 A0A3B0J517 A0A1A9VAQ6 B4LDZ7 A0A0L0CRG4 T1PI12 A0A1B0CA01 A0A1B0GNL3 A0A1W4WS98 A0A1L8DY41 A0A182HH43 A0A1W4X375 D2A2T8 A0A1J1J6T8 A0A1Y1KH18 A0A182TRT0 A0A2J7QBB1 A0A1W4X1X2 E0VXZ2 N6USQ5 A0A1B6CWK0 U4USR9 A0A067R5K5 A0A2M4ARK8 A0A1J1IG59 A0A154NW13 T1HWS6 A0A0A9YIY7 A0A023F1G3 A0A2P8ZB90 A0A087ZX11 A0A423T4A1 A0A0N0BHP6 E2AAK6 E2BT41 A0A0L7R635 A0A2A3EKM2 A0A026W0G8 A0A232ERU2 A0A151WSR4 A0A310SKT0 A0A195BVE3 A0A3Q0ISL3 A0A195FPW6 A0A3L8DDG7 A0A195CAV7 A0A2P2I5D9 A0A158NV05 F4WJF9
A0A182FKP1 A0A2M4APL5 W5JDI0 A0A182NPQ1 A0A182QDB5 A0A182P6X8 A0A182RIG0 A0A182YL43 A0A182SZY8 A0A182WEC7 A0A182JZP8 A0A182M6N3 A0A084VIP0 A0A1Q3FVR8 A0A1Q3FW47 A0A182INR1 A0A182G5W6 A0A2Y9D3G6 A0A182V4Y4 A0A182L4K3 Q7PZ51 A0A1I8P9M2 A0A336K7Y1 B4N727 M9PFE4 Q9VUW9 A0A1A9XT26 A0A1B0B966 Q29D93 B4H7T3 W8BBQ0 B4ITV5 B3M9T9 B3NDQ1 A0A034VE00 A0A1A9WGH9 B4QLS4 A0A1B0FAV5 B4KYH8 A0A0K8UBM9 A0A1W4VAE9 A0A0K8U1C2 A0A0K8UW18 B4J3N8 A0A0M3QWV8 A0A1B0AIE9 A0A3B0J517 A0A1A9VAQ6 B4LDZ7 A0A0L0CRG4 T1PI12 A0A1B0CA01 A0A1B0GNL3 A0A1W4WS98 A0A1L8DY41 A0A182HH43 A0A1W4X375 D2A2T8 A0A1J1J6T8 A0A1Y1KH18 A0A182TRT0 A0A2J7QBB1 A0A1W4X1X2 E0VXZ2 N6USQ5 A0A1B6CWK0 U4USR9 A0A067R5K5 A0A2M4ARK8 A0A1J1IG59 A0A154NW13 T1HWS6 A0A0A9YIY7 A0A023F1G3 A0A2P8ZB90 A0A087ZX11 A0A423T4A1 A0A0N0BHP6 E2AAK6 E2BT41 A0A0L7R635 A0A2A3EKM2 A0A026W0G8 A0A232ERU2 A0A151WSR4 A0A310SKT0 A0A195BVE3 A0A3Q0ISL3 A0A195FPW6 A0A3L8DDG7 A0A195CAV7 A0A2P2I5D9 A0A158NV05 F4WJF9
EC Number
2.3.1.225
Pubmed
19121390
26354079
28756777
20920257
23761445
25244985
+ More
24438588 26483478 20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 18158335 18032660 18719403 20599894 15632085 24495485 17550304 25348373 22936249 26108605 25315136 18362917 19820115 28004739 20566863 23537049 24845553 25401762 25474469 29403074 20798317 24508170 28648823 30249741 21347285 21719571
24438588 26483478 20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 18158335 18032660 18719403 20599894 15632085 24495485 17550304 25348373 22936249 26108605 25315136 18362917 19820115 28004739 20566863 23537049 24845553 25401762 25474469 29403074 20798317 24508170 28648823 30249741 21347285 21719571
EMBL
BABH01000003
ODYU01003924
SOQ43287.1
NWSH01001493
PCG71089.1
KQ460297
+ More
KPJ16263.1 KZ150387 PZC71058.1 KQ459439 KPJ01096.1 GGFK01009227 MBW42548.1 ADMH02001696 ETN61393.1 AXCN02001661 AXCM01000779 ATLV01013376 KE524854 KFB37834.1 GFDL01003407 JAV31638.1 GFDL01003402 JAV31643.1 JXUM01044388 KQ561398 KXJ78710.1 AAAB01008986 EAA00049.3 UFQS01000115 UFQT01000115 SSW99922.1 SSX20302.1 CH964168 EDW80166.2 AE014296 AGB94603.1 AY094779 AAM11132.1 JXJN01010285 CH379070 EAL30521.2 CH479219 EDW34723.1 GAMC01007900 JAB98655.1 CH891734 EDW99818.2 CH902618 EDV40130.2 CH954178 EDV52184.2 GAKP01019189 JAC39763.1 CM000363 CM002912 EDX10623.1 KMY99892.1 CCAG010022451 CH933809 EDW18789.2 GDHF01028232 JAI24082.1 GDHF01032179 JAI20135.1 GDHF01021759 JAI30555.1 CH916366 EDV97269.1 CP012525 ALC44789.1 OUUW01000002 SPP76797.1 CH940647 EDW70040.2 JRES01000005 KNC34955.1 KA648324 AFP62953.1 AJWK01003138 AJVK01029539 GFDF01002750 JAV11334.1 APCN01002385 KQ971339 EFA01463.1 CVRI01000074 CRL08133.1 GEZM01086196 JAV59681.1 NEVH01016301 PNF25878.1 DS235842 EEB18248.1 APGK01017930 APGK01017931 APGK01017932 KB740060 ENN81782.1 GEDC01019481 JAS17817.1 KB632433 ERL95568.1 KK852729 KDR17567.1 GGFK01010083 MBW43404.1 CVRI01000047 CRK98524.1 KQ434772 KZC03869.1 ACPB03005829 ACPB03005830 GBHO01010557 JAG33047.1 GBBI01003350 JAC15362.1 PYGN01000117 PSN53756.1 QCYY01002305 ROT71356.1 KQ435746 KOX76561.1 GL438128 EFN69553.1 GL450325 EFN81171.1 KQ414648 KOC66289.1 KZ288219 PBC32315.1 KK107519 EZA49450.1 NNAY01002550 OXU21075.1 KQ982769 KYQ50878.1 KQ761502 OAD57473.1 KQ976401 KYM92594.1 KQ981335 KYN42635.1 QOIP01000009 RLU18497.1 KQ978023 KYM97942.1 IACF01003631 LAB69242.1 ADTU01026871 GL888182 EGI65737.1
KPJ16263.1 KZ150387 PZC71058.1 KQ459439 KPJ01096.1 GGFK01009227 MBW42548.1 ADMH02001696 ETN61393.1 AXCN02001661 AXCM01000779 ATLV01013376 KE524854 KFB37834.1 GFDL01003407 JAV31638.1 GFDL01003402 JAV31643.1 JXUM01044388 KQ561398 KXJ78710.1 AAAB01008986 EAA00049.3 UFQS01000115 UFQT01000115 SSW99922.1 SSX20302.1 CH964168 EDW80166.2 AE014296 AGB94603.1 AY094779 AAM11132.1 JXJN01010285 CH379070 EAL30521.2 CH479219 EDW34723.1 GAMC01007900 JAB98655.1 CH891734 EDW99818.2 CH902618 EDV40130.2 CH954178 EDV52184.2 GAKP01019189 JAC39763.1 CM000363 CM002912 EDX10623.1 KMY99892.1 CCAG010022451 CH933809 EDW18789.2 GDHF01028232 JAI24082.1 GDHF01032179 JAI20135.1 GDHF01021759 JAI30555.1 CH916366 EDV97269.1 CP012525 ALC44789.1 OUUW01000002 SPP76797.1 CH940647 EDW70040.2 JRES01000005 KNC34955.1 KA648324 AFP62953.1 AJWK01003138 AJVK01029539 GFDF01002750 JAV11334.1 APCN01002385 KQ971339 EFA01463.1 CVRI01000074 CRL08133.1 GEZM01086196 JAV59681.1 NEVH01016301 PNF25878.1 DS235842 EEB18248.1 APGK01017930 APGK01017931 APGK01017932 KB740060 ENN81782.1 GEDC01019481 JAS17817.1 KB632433 ERL95568.1 KK852729 KDR17567.1 GGFK01010083 MBW43404.1 CVRI01000047 CRK98524.1 KQ434772 KZC03869.1 ACPB03005829 ACPB03005830 GBHO01010557 JAG33047.1 GBBI01003350 JAC15362.1 PYGN01000117 PSN53756.1 QCYY01002305 ROT71356.1 KQ435746 KOX76561.1 GL438128 EFN69553.1 GL450325 EFN81171.1 KQ414648 KOC66289.1 KZ288219 PBC32315.1 KK107519 EZA49450.1 NNAY01002550 OXU21075.1 KQ982769 KYQ50878.1 KQ761502 OAD57473.1 KQ976401 KYM92594.1 KQ981335 KYN42635.1 QOIP01000009 RLU18497.1 KQ978023 KYM97942.1 IACF01003631 LAB69242.1 ADTU01026871 GL888182 EGI65737.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000069272
UP000000673
+ More
UP000075884 UP000075886 UP000075885 UP000075900 UP000076408 UP000075901 UP000075920 UP000075881 UP000075883 UP000030765 UP000075880 UP000069940 UP000249989 UP000076407 UP000075903 UP000075882 UP000007062 UP000095300 UP000007798 UP000000803 UP000092443 UP000092460 UP000001819 UP000008744 UP000002282 UP000007801 UP000008711 UP000091820 UP000000304 UP000092444 UP000009192 UP000192221 UP000001070 UP000092553 UP000092445 UP000268350 UP000078200 UP000008792 UP000037069 UP000095301 UP000092461 UP000092462 UP000192223 UP000075840 UP000007266 UP000183832 UP000075902 UP000235965 UP000009046 UP000019118 UP000030742 UP000027135 UP000076502 UP000015103 UP000245037 UP000005203 UP000283509 UP000053105 UP000000311 UP000008237 UP000053825 UP000242457 UP000053097 UP000215335 UP000075809 UP000078540 UP000079169 UP000078541 UP000279307 UP000078542 UP000005205 UP000007755
UP000075884 UP000075886 UP000075885 UP000075900 UP000076408 UP000075901 UP000075920 UP000075881 UP000075883 UP000030765 UP000075880 UP000069940 UP000249989 UP000076407 UP000075903 UP000075882 UP000007062 UP000095300 UP000007798 UP000000803 UP000092443 UP000092460 UP000001819 UP000008744 UP000002282 UP000007801 UP000008711 UP000091820 UP000000304 UP000092444 UP000009192 UP000192221 UP000001070 UP000092553 UP000092445 UP000268350 UP000078200 UP000008792 UP000037069 UP000095301 UP000092461 UP000092462 UP000192223 UP000075840 UP000007266 UP000183832 UP000075902 UP000235965 UP000009046 UP000019118 UP000030742 UP000027135 UP000076502 UP000015103 UP000245037 UP000005203 UP000283509 UP000053105 UP000000311 UP000008237 UP000053825 UP000242457 UP000053097 UP000215335 UP000075809 UP000078540 UP000079169 UP000078541 UP000279307 UP000078542 UP000005205 UP000007755
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9IV03
A0A2H1VR64
A0A2A4JGZ5
A0A194REK0
A0A2W1B5J2
A0A194QCD4
+ More
A0A182FKP1 A0A2M4APL5 W5JDI0 A0A182NPQ1 A0A182QDB5 A0A182P6X8 A0A182RIG0 A0A182YL43 A0A182SZY8 A0A182WEC7 A0A182JZP8 A0A182M6N3 A0A084VIP0 A0A1Q3FVR8 A0A1Q3FW47 A0A182INR1 A0A182G5W6 A0A2Y9D3G6 A0A182V4Y4 A0A182L4K3 Q7PZ51 A0A1I8P9M2 A0A336K7Y1 B4N727 M9PFE4 Q9VUW9 A0A1A9XT26 A0A1B0B966 Q29D93 B4H7T3 W8BBQ0 B4ITV5 B3M9T9 B3NDQ1 A0A034VE00 A0A1A9WGH9 B4QLS4 A0A1B0FAV5 B4KYH8 A0A0K8UBM9 A0A1W4VAE9 A0A0K8U1C2 A0A0K8UW18 B4J3N8 A0A0M3QWV8 A0A1B0AIE9 A0A3B0J517 A0A1A9VAQ6 B4LDZ7 A0A0L0CRG4 T1PI12 A0A1B0CA01 A0A1B0GNL3 A0A1W4WS98 A0A1L8DY41 A0A182HH43 A0A1W4X375 D2A2T8 A0A1J1J6T8 A0A1Y1KH18 A0A182TRT0 A0A2J7QBB1 A0A1W4X1X2 E0VXZ2 N6USQ5 A0A1B6CWK0 U4USR9 A0A067R5K5 A0A2M4ARK8 A0A1J1IG59 A0A154NW13 T1HWS6 A0A0A9YIY7 A0A023F1G3 A0A2P8ZB90 A0A087ZX11 A0A423T4A1 A0A0N0BHP6 E2AAK6 E2BT41 A0A0L7R635 A0A2A3EKM2 A0A026W0G8 A0A232ERU2 A0A151WSR4 A0A310SKT0 A0A195BVE3 A0A3Q0ISL3 A0A195FPW6 A0A3L8DDG7 A0A195CAV7 A0A2P2I5D9 A0A158NV05 F4WJF9
A0A182FKP1 A0A2M4APL5 W5JDI0 A0A182NPQ1 A0A182QDB5 A0A182P6X8 A0A182RIG0 A0A182YL43 A0A182SZY8 A0A182WEC7 A0A182JZP8 A0A182M6N3 A0A084VIP0 A0A1Q3FVR8 A0A1Q3FW47 A0A182INR1 A0A182G5W6 A0A2Y9D3G6 A0A182V4Y4 A0A182L4K3 Q7PZ51 A0A1I8P9M2 A0A336K7Y1 B4N727 M9PFE4 Q9VUW9 A0A1A9XT26 A0A1B0B966 Q29D93 B4H7T3 W8BBQ0 B4ITV5 B3M9T9 B3NDQ1 A0A034VE00 A0A1A9WGH9 B4QLS4 A0A1B0FAV5 B4KYH8 A0A0K8UBM9 A0A1W4VAE9 A0A0K8U1C2 A0A0K8UW18 B4J3N8 A0A0M3QWV8 A0A1B0AIE9 A0A3B0J517 A0A1A9VAQ6 B4LDZ7 A0A0L0CRG4 T1PI12 A0A1B0CA01 A0A1B0GNL3 A0A1W4WS98 A0A1L8DY41 A0A182HH43 A0A1W4X375 D2A2T8 A0A1J1J6T8 A0A1Y1KH18 A0A182TRT0 A0A2J7QBB1 A0A1W4X1X2 E0VXZ2 N6USQ5 A0A1B6CWK0 U4USR9 A0A067R5K5 A0A2M4ARK8 A0A1J1IG59 A0A154NW13 T1HWS6 A0A0A9YIY7 A0A023F1G3 A0A2P8ZB90 A0A087ZX11 A0A423T4A1 A0A0N0BHP6 E2AAK6 E2BT41 A0A0L7R635 A0A2A3EKM2 A0A026W0G8 A0A232ERU2 A0A151WSR4 A0A310SKT0 A0A195BVE3 A0A3Q0ISL3 A0A195FPW6 A0A3L8DDG7 A0A195CAV7 A0A2P2I5D9 A0A158NV05 F4WJF9
PDB
5W7I
E-value=6.15963e-75,
Score=716
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
Cell junction Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
Synapse Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
Presynaptic cell membrane Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
Cell junction Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
Synapse Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
Presynaptic cell membrane Localizes to the Golgi apparatus in non-neuronal cells and located primarily in presynaptic terminals in neurons. With evidence from 5 publications.
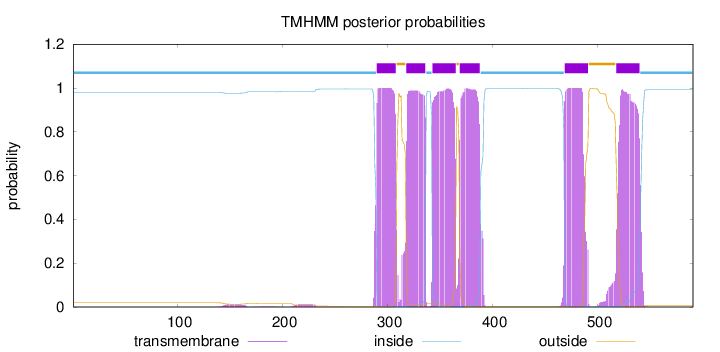
Length:
591
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
126.16845
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98083
inside
1 - 289
TMhelix
290 - 308
outside
309 - 317
TMhelix
318 - 336
inside
337 - 342
TMhelix
343 - 365
outside
366 - 368
TMhelix
369 - 388
inside
389 - 468
TMhelix
469 - 491
outside
492 - 517
TMhelix
518 - 540
inside
541 - 591
Population Genetic Test Statistics
Pi
5.053229
Theta
6.595845
Tajima's D
-1.084509
CLR
0.101596
CSRT
0.11704414779261
Interpretation
Uncertain
