Gene
KWMTBOMO08147
Pre Gene Modal
BGIBMGA001082
Annotation
PREDICTED:_MOB_kinase_activator-like_2_isoform_X3_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.91
Sequence
CDS
ATGATATCCTCCATAATTTGCAAACAACAAAATAGCGGCGGATGCGGTGGCCCGGTGTCGTTGGAGTCGGAGTGGAGGAGTGCGGGCCGCGGGAGGGGAAGAGCGCGCGCGGCGCGTACACTCGGCGCCTCTCAGTCGTCGCTCGTCGCCGCGATGTGTCACGCCGTCGTGCTCGGTGTGTTGTTCTCATTGCCCTCCTACCTGTACTCGGCGGTCCCGTCCAGGAAGGCTCGTCGGAAGGAGCGAGAAGGGGAGCCGGCCGGAGACCCGAAGCTGTACCTGCAGGAGGCGCTGCTCGAGCGGAAGCTCCCCGAACTGGACATGCGCCAGCTCGTCGACCTGCCCCACGGACTCGACTACAACGAATGGCTTGCTTCGCACACGCTGGCGCTATTCGACCACGTGAACCTGGTGTACGGCACGGTGTCGGAGTTCTGCACGGCGGCCACGTGCCCCGACATGACGGGCCCGGGCGGGCGCAGCTACGCGTGGTACGACGAGCGCGGCAAGAAGTCCCGCGTCGCCGCGCCGCAGTACGTCGACTACGTCATGACCTACACGCAGAAGACTGTCAACGATGAGACCGTCTTCCCGACTAAATATGCGAACGAGTTCCCCGCGCAGTTCGAGGGCGTGGTGCGGCGCGTGGTGCGGCTGCTGTTCCACGTGGTGGCGCACATGTACGCCGCGCACTTCCGCGAGCTCGCGCTGCTGCGGCTGCACGCGCACCTGCACCTCACCTTCGCGCACCTCACCGCGCTCGACCGCCGCTACGCGCTGCTCGACCACAAGGAGACCGACGTGCTGCGCGACCTCGAGGTGGCGCTGCGCCTGCGCGAGCCGCCCTCCCCGCGCGCCGCCCCCGCCGCGCCCGCCCCGCCCCCCGCCGCGCCCGCACCCCCCGCCGCGCCCGCACCGCCCGCCGCGCCCGCGCCGCCCCCCGCCGCCACCGCGTGA
Protein
MISSIICKQQNSGGCGGPVSLESEWRSAGRGRGRARAARTLGASQSSLVAAMCHAVVLGVLFSLPSYLYSAVPSRKARRKEREGEPAGDPKLYLQEALLERKLPELDMRQLVDLPHGLDYNEWLASHTLALFDHVNLVYGTVSEFCTAATCPDMTGPGGRSYAWYDERGKKSRVAAPQYVDYVMTYTQKTVNDETVFPTKYANEFPAQFEGVVRRVVRLLFHVVAHMYAAHFRELALLRLHAHLHLTFAHLTALDRRYALLDHKETDVLRDLEVALRLREPPSPRAAPAAPAPPPAAPAPPAAPAPPAAPAPPPAATA
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF03637 Mob1_phocein
SUPFAM
SSF101152
SSF101152
Gene 3D
ProteinModelPortal
PDB
5BRK
E-value=6.67002e-25,
Score=282
Ontologies
GO
PANTHER
Topology
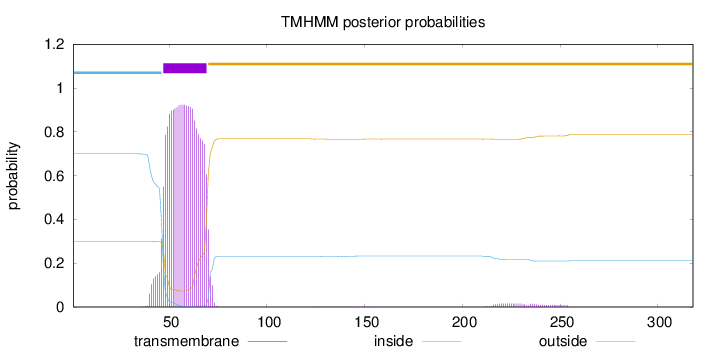
Length:
318
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.54553
Exp number, first 60 AAs:
13.25074
Total prob of N-in:
0.69948
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 69
outside
70 - 318
Population Genetic Test Statistics
Pi
2.642693
Theta
6.886002
Tajima's D
-1.707413
CLR
0.132617
CSRT
0.0300984950752462
Interpretation
Uncertain
