Gene
KWMTBOMO08144
Pre Gene Modal
BGIBMGA001079
Annotation
receptor_protein-tyrosine_phosphatase_10d_[Danaus_plexippus]
Full name
Tyrosine-protein phosphatase 10D
Alternative Name
Receptor-linked protein-tyrosine phosphatase 10D
Location in the cell
Mitochondrial Reliability : 1.285 Nuclear Reliability : 1.042 PlasmaMembrane Reliability : 1.203
Sequence
CDS
ATGACGTGGAAAAGGTTGAACTTGCAAGCGACTGGAGGATGCGCACGCGCTGCGGCCCTGGCACTGCTTCTAGTTATCCTTGCGTGCATGACGAGGTGCGGCTCGTGTGCCGACCTGGTGATCGAGATCCCGGGCTCCGGCGCGGGCGAGGATGGAGACAGCGTGAGCGGCTTGTACCGGCTAGACTACCGGCCGCCCGAGGGCACGCCCGCCCCGAACCACACCGTCCCCGCGCGCGCCTCTACCATTAACTTTCAAGGCCTGCCCGGCACCAAGTATCATTTCATGCTCTACTACTCCAACTCTACCTTCCCTGATTTGCTCACCTGGAACCAAACGATAATTACGGCGCCGGAGCCGCCAACGAACTTGACGGTGACCTTGGGCCGCAACAAGCAAGCCACCATATCCTGGTCGCCACCCACCCAGGGTAACTATACGGGCTTTCGTTTTAAGGTGATCCCGCTGACGGAGCGCGCGGAGGGCGGCCAGCGCAACATCACGGTGGAGCGCGGCAACGACACGCACGTGCTGCGCGACCTGTTCCCCGGCGCCACCTACCAGCTGCACGCCTTCACGCTGCTGCACGACAAGGAGAGCGCCGCCTACGCCTCGCGCAACTTCACCACAAAGCCTAACACGCCCGGCAAGTTCATCGTGTGGTTCCGCAACGAGACCACGCTGCTGGTGCTGTGGCAGCCGCCCTACCCGCCCGGCGCCTACACGCACTACAAGGTGTCGATCGAGCCGCCCGATGCTCGCGACTCGGTGCTATACGTGGAGAAGGAGGGCGAGCCGCCCGGCCCCGCCCAGGCCGCCTTCAAAGGTCTGGTGCCCGGGCGGGCCTACAACATCTCGGTGCAGACCGTCTCCGAGCCCGAGATCTCGGCGCCCACCACCGCCAAGTACCGCACCGTGCCGCTGCGGCCGCGCAACGTGACCGCGCCGCCCGACGACCTGCGCGAGACTGCCTTCCGCGTCACGTGGCTGCCGCCCCTGGAGGAGAGCGAGTTCGAGAAGTACCAGGTGTCGGTGTCCGGGGGCGGGGCGGCCCGCCGGCTGAGCCCGCTGCTGCTGGGCCGCGACGAGGAGCCGTCCTGCTGGTTCAGGCGGCTGGACCCGGGCGCGCACTACTCGCTCACCGTCAAGACCATGGCCGGGAAGGTGACCTCCTGGCCCGCCGCCCTCGACCTCACGCTCAAGCCGCTGCCGGTGCGGCGCCTGCTGGTGAGCTCGGCGGAGGGCGGGCTGGGCGTGGCGTGGCAGCCCGCCGACGGCAGCGCGCAGGACGAGTACAAGGTCTCCTACCACGAGGCGGGCCCCTCGCGCGACGACAGCAACACGCTGACCACCACCGACACCAACGTGACGCTGGACGCGCTGCTGCCCGGCCGCAACTACACCGTGTCCGTGCGCGCGCTGTCCCGCGCCGTGGAGTCCAACGAGACCTGGGGCTGGGGCGCCACGCGGCCGCTGGCCCCCGTGGTGGTGCGCGTGGAGGCCGCCGCCGCCGCCCTGCGCCTGCACTGGCGCTCCGACGTCAACTCGCGCCAGACGCGCTACGTGCTGCGCTACCGGGCGCGCGTGCCGGAGCCCGGGCCGCCGGCCGAGTGGCGCCAGCTGCGGACCGAGGACACCAGCGCGCTGCTGGCCGACCTGTTCCCGGGCGCCGTGTACGAGATCGAGCTCGCCGCCATGAGCCACGACCTGCCGAGCGAGGCGCACCGCATGCTCAAGCCCGTGCGGCCGCTGGCTGCCACCCGCCTGTGGGTGGAGCGCGCCTCCTCCAACAGCGCGCTGGTGCAGTGGCGCGGCCCGCCGCCCGCCCGCAGCGTGCTGGGCCACTACGCGCTGCGCTACCGCACGCTGTCCGCCGCGCGCTTCACCGAGCTGGACCTGCTGCCGCCCGACGCCGCGTCCGCCGAGATCACCAACATGACGCACGGGGAGCGCTACGTCATCCAGCTGGACACGGTCAGCGAGCCCGGCGACGGCGGCGAGGTGGTGGAGAGCGGGCAGCCGCTGTACGCCGAGCACACCATCCGTCCCAACCCGGTGTCCGAGGTGGCGCAGCTGGTGGGCACCCGCAACGTCACGCTCGAATGGCTGAACGGCTCCGGACGCGTCGACCGGTACGAGGTGCGCTGGTGGCCCACGGACCACGCCGAGCTGGCCGGGCTGGAGGGCACCGACGAGGACTCTGCGCGGGGCGCGGCGGGCGGCGAGCGGGCCCGCAACCTCACGGCGCCGCCGGCGCTGCTGGCGGGGCTGGCGCCGGGCGTGGGCTACACGCTGACCCTGGCCGCGCACTCCTACAACCTCACCAGCGACCTGCTCACCATACACACGCGAACACGTCCGCTGATCCTGTCCGAGATGACCATCGTGAACGAGCCCAGCGACGACGAGGAGGCGCCGCCCGGCATCATCGTTCTGTTCACGCCGACGCCGAGCTCCGCCACGCGCTTCGACCAGTACCGCTTCCTGCTGGAGGGCGCCGCCGACGAGCCGCCGCGCGACGCCGCCCTGCCCGCGCAGCCCGCGCCCCCCGCGCCGCGCCGCGTGCACTGGCCCGGCCTCGTGCCCGGCCGCCTCTATGACGTCACCATGTGGACGCTCAGCGCCAACGTGACCTCGCACCCCGTGCAGCGCCAGACCAGGCTGCACCCGCGGCCCGTGCGCGCGCTCAACACGACGCGCGTGGGTGCGCGGTTCGTGACGCTGACCTGGGAGCGGCCTGGGGGCGACCACACTGACTTCGAGCTCCAGTATTTCGCCGACGAACAAGTCCGCACCCTGGTCACCGACCGGCTGTCCATCAACGCGACCGACTTGGAGCCGTACACGTCGTACGATTTCACCGTCAGGGTGCGGTCCGGGGACGGCGGCGGCCTGCGCATGGTGTCTCCGGAGCGCAGCGTGGCCGTCCGCACCGCCGAGGACGTGCCCGCCGCGCCCGCCGACTTCCGGCCGGCCGACGCGCGGCCCAGCTCCCTCACCATGGCGTGGCGGCTGTCGGCGGCGCAGGCCCACGGACTGCTGCGCCGCTACGTGCTCACGTACGCCCCGCACGGCCGCCCCGAGCTCACGCGCTCGCTGGAGCTGGCGGCGGAGGCCACGCGCGTCACGGTGGAGGGCTTGTCGCCCGGGGTCACGTACGAACTGAGTCTGCACGGGGAGACGGGCGCCGGCCCGGGGGCGGCCGCGCACTGGACGCAGCACATGGCCATCGGCGCGCCGCCCGCCCCGCCGCCGGACGTGGCGCCGGCCGCCGTGCGCGCTACCACCACGACGGTCGTGGTGCGGTTCCGCTCTGACTACTTCTCGGCAGTCAACGGCAACGTGACCGCGTACACGCTGGTGCTGGCGCAGGAGAACCGCTCGGAGACGCCGGCGCGGCTGCCGTCCTGGCGCGACGTGCACGCGCTGCCGGTGTGGCCGCCCTACCAGGTGACGGAGCCCTACTACCCGTTCCTGTCGTCGCCGGTGGAGGAGTTCACGCTGGGTTCGGAGCGGTGCGAGGGAGGGGGCCGGGCATACTGCAACGGTCCGCTGAAGGCAGGCACGCGATACTTCTTGAAGCTGCGCGCCTTCACCGCGCCCGACAAGTTCGCCGACACCGCCTACGTCGCCGTGGCCACGGAGGTGGACAACACGGCGTGGATCATCGGCGGCAGCGTGTGCGGGCTGGCGGTGGCGGGCGCGGCGCTGGCGGCGCTGGCTGCCTCCCGCCGCCGCCGCCGGGCCGCGCGCGCACCGGCCGCGCCCGCGCCGCGCCGCACCTCCCGCCCCGTGCGCGTGGCGGACTTCATCGACCACTACCGAGCCATGTCCGCCGACTCCGACTTCAAGTTCAGCGAGGAGTTCGAGGAGCTGAAGGACGTGGGCCGCGAGCAGCCGTGCACGGCGGCCGACCTGCCCGTGAACCGTCCCAAGAACCGGTTCACCAACATCCTGCCGTACGACCACTCGCGCTACAAGCTGCAGCCCGTCGACGACGAGGAGGGCTCCGACTACATCAACGCCAACTACCTGCCCGGGTACAACTCGCCCCGGGAGTTCATCGTGACGCAGGGCCCGCTGCACTGCACCCGGGACGACTTCTGGCGCATGTGCTGGGAGTCGGGCTCGCGCGCCGTCGTGATGCTGACTCGCTGCGTGGAGAAGGGCCGCGAGAAGTGCGACCGGTACTGGCCCTACGACACGCGGCCCGTGTACTACGGGGACATCGCCGTGACCGCGCTGAACGAGAGCCGCTACCCCGACTGGACCGTCACGGAGCTGCTGGTGAGCCGCGGCAAGGAGCAGCGCGTGCTGCTGCACTTCCACTTCACCACGTGGCCAGACTTCGGCGTGCCCGAGCCGCCCACCACGCTGGCGCGCTTCGTGCGGGCCTTCCGCGAGCGCTGCCCGGCCGACGGCCGCCCCGTGGTGGTGCACTGCAGCGCCGGCGTGGGCCGCTCCGGCACCTTCATCACGCTGGACACGGCGCTGCAGCAGCTGGCGGCGCGCGACGACTACGTCGACATCTTCGGCATGGTGTACAACATGCGCCGCGAGCGCGTGGCCATGGTGCAGTCGGAGCAGCAGTACATCTGCATCCACCAGTGCCTCGTGGCCGTGCTGGAGGGCCAGGACCTGACGCCCCCGCACAACAACAACCACCACCACAACCCGGCCTTCGAAGACGACGAGGGCATCGCGGAGTCCGGGATGTGA
Protein
MTWKRLNLQATGGCARAAALALLLVILACMTRCGSCADLVIEIPGSGAGEDGDSVSGLYRLDYRPPEGTPAPNHTVPARASTINFQGLPGTKYHFMLYYSNSTFPDLLTWNQTIITAPEPPTNLTVTLGRNKQATISWSPPTQGNYTGFRFKVIPLTERAEGGQRNITVERGNDTHVLRDLFPGATYQLHAFTLLHDKESAAYASRNFTTKPNTPGKFIVWFRNETTLLVLWQPPYPPGAYTHYKVSIEPPDARDSVLYVEKEGEPPGPAQAAFKGLVPGRAYNISVQTVSEPEISAPTTAKYRTVPLRPRNVTAPPDDLRETAFRVTWLPPLEESEFEKYQVSVSGGGAARRLSPLLLGRDEEPSCWFRRLDPGAHYSLTVKTMAGKVTSWPAALDLTLKPLPVRRLLVSSAEGGLGVAWQPADGSAQDEYKVSYHEAGPSRDDSNTLTTTDTNVTLDALLPGRNYTVSVRALSRAVESNETWGWGATRPLAPVVVRVEAAAAALRLHWRSDVNSRQTRYVLRYRARVPEPGPPAEWRQLRTEDTSALLADLFPGAVYEIELAAMSHDLPSEAHRMLKPVRPLAATRLWVERASSNSALVQWRGPPPARSVLGHYALRYRTLSAARFTELDLLPPDAASAEITNMTHGERYVIQLDTVSEPGDGGEVVESGQPLYAEHTIRPNPVSEVAQLVGTRNVTLEWLNGSGRVDRYEVRWWPTDHAELAGLEGTDEDSARGAAGGERARNLTAPPALLAGLAPGVGYTLTLAAHSYNLTSDLLTIHTRTRPLILSEMTIVNEPSDDEEAPPGIIVLFTPTPSSATRFDQYRFLLEGAADEPPRDAALPAQPAPPAPRRVHWPGLVPGRLYDVTMWTLSANVTSHPVQRQTRLHPRPVRALNTTRVGARFVTLTWERPGGDHTDFELQYFADEQVRTLVTDRLSINATDLEPYTSYDFTVRVRSGDGGGLRMVSPERSVAVRTAEDVPAAPADFRPADARPSSLTMAWRLSAAQAHGLLRRYVLTYAPHGRPELTRSLELAAEATRVTVEGLSPGVTYELSLHGETGAGPGAAAHWTQHMAIGAPPAPPPDVAPAAVRATTTTVVVRFRSDYFSAVNGNVTAYTLVLAQENRSETPARLPSWRDVHALPVWPPYQVTEPYYPFLSSPVEEFTLGSERCEGGGRAYCNGPLKAGTRYFLKLRAFTAPDKFADTAYVAVATEVDNTAWIIGGSVCGLAVAGAALAALAASRRRRRAARAPAAPAPRRTSRPVRVADFIDHYRAMSADSDFKFSEEFEELKDVGREQPCTAADLPVNRPKNRFTNILPYDHSRYKLQPVDDEEGSDYINANYLPGYNSPREFIVTQGPLHCTRDDFWRMCWESGSRAVVMLTRCVEKGREKCDRYWPYDTRPVYYGDIAVTALNESRYPDWTVTELLVSRGKEQRVLLHFHFTTWPDFGVPEPPTTLARFVRAFRERCPADGRPVVVHCSAGVGRSGTFITLDTALQQLAARDDYVDIFGMVYNMRRERVAMVQSEQQYICIHQCLVAVLEGQDLTPPHNNNHHHNPAFEDDEGIAESGM
Summary
Description
May have a role in axon outgrowth and guidance.
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Miscellaneous
This protein is translated by readthrough of a stop codon. Readthrough of the terminator codon TAG occurs between the codons for Arg-1631 and His-1633. There is currently no sequence that provides the identity of residue 1632.
Similarity
Belongs to the protein-tyrosine phosphatase family. Receptor class subfamily.
Keywords
3D-structure
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Glycoprotein
Hydrolase
Membrane
Neurogenesis
Protein phosphatase
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Tyrosine-protein phosphatase 10D
splice variant In isoform H.
splice variant In isoform H.
Uniprot
A0A212EVL1
A0A1S4FVE7
Q16N57
A0A2C9GXN6
A0A1Q3FEP0
A0A1Q3FEM2
+ More
Q7PQL0 A0A2C9GPF3 A0A2C9H7N3 A0A2M3ZFK9 A0A2M4A1D2 W4VRK2 A0A182Y5C9 A0A2M4CPV4 A0A1S4GKI9 A0A1S4GL07 A0A2M4CNN4 A0A182PD62 A0A182V608 F5HL66 A0A2M4B965 A0A2M4B8X5 A0A2C9H7M9 A0A2C9GPF4 A0A182XG36 A0A2C9GPF8 A0A182W196 A0A182RX49 B0WFI2 A0A182QZU5 A0A3L8DBY3 W5JMK0 A0A182MFC8 A0A182FPT1 A0A182GX55 A0A182NCZ2 A0A182K559 A0A2H1WIJ0 A0A2M4CPH8 E2AHA6 A0A182ISY5 T1PDC5 W8AUE6 A0A0K8UAK8 A0A3B0KR32 E9IF67 A0A182L725 D2A2C1 A0A146LQN5 A0A0A1WVW3 A0A0P4VYC9 A0A1W4VJU9 A0A1B0G7C0 P35992-2 A0A158NAA8 A0A1B0AMN8 K7IYR7 A0A0L0BUR2 A0A1A9X001 B3MXI6 A0A1I8NIH2 A0A023F268 B4NBT8 F4WN75 A0A310STH1 A0A1L8DLG1 Q29IE5 A0A1L8DLD8 A0A1B0AHB4 A0A0K8UU60 A0A1W4VJ84 A0A139WIH4 A0A1I8P7V3 B4Q1S6 A0A0C9R9Q6 A0A0A1X0Z4 P35992 A0A1W4VKM0 A0A232FFA9 A0A0K8U159 P35992-1 A0A1A9UY21 B4JK57 B3NUA6 A0A1B0DK97 B4IK45 A0A1J1HJR5 A0A1J1HJB1 A0A1W4WAG1 A0A1W4V7G9 A0A0C9R0K3 P35992-6 A0A1J1HIC1 A0A026WK83 B4L685 U4UAM5 A0A336MWI6 A0A336MU49 A0A2J7REX4 A0A2R7WCI1
Q7PQL0 A0A2C9GPF3 A0A2C9H7N3 A0A2M3ZFK9 A0A2M4A1D2 W4VRK2 A0A182Y5C9 A0A2M4CPV4 A0A1S4GKI9 A0A1S4GL07 A0A2M4CNN4 A0A182PD62 A0A182V608 F5HL66 A0A2M4B965 A0A2M4B8X5 A0A2C9H7M9 A0A2C9GPF4 A0A182XG36 A0A2C9GPF8 A0A182W196 A0A182RX49 B0WFI2 A0A182QZU5 A0A3L8DBY3 W5JMK0 A0A182MFC8 A0A182FPT1 A0A182GX55 A0A182NCZ2 A0A182K559 A0A2H1WIJ0 A0A2M4CPH8 E2AHA6 A0A182ISY5 T1PDC5 W8AUE6 A0A0K8UAK8 A0A3B0KR32 E9IF67 A0A182L725 D2A2C1 A0A146LQN5 A0A0A1WVW3 A0A0P4VYC9 A0A1W4VJU9 A0A1B0G7C0 P35992-2 A0A158NAA8 A0A1B0AMN8 K7IYR7 A0A0L0BUR2 A0A1A9X001 B3MXI6 A0A1I8NIH2 A0A023F268 B4NBT8 F4WN75 A0A310STH1 A0A1L8DLG1 Q29IE5 A0A1L8DLD8 A0A1B0AHB4 A0A0K8UU60 A0A1W4VJ84 A0A139WIH4 A0A1I8P7V3 B4Q1S6 A0A0C9R9Q6 A0A0A1X0Z4 P35992 A0A1W4VKM0 A0A232FFA9 A0A0K8U159 P35992-1 A0A1A9UY21 B4JK57 B3NUA6 A0A1B0DK97 B4IK45 A0A1J1HJR5 A0A1J1HJB1 A0A1W4WAG1 A0A1W4V7G9 A0A0C9R0K3 P35992-6 A0A1J1HIC1 A0A026WK83 B4L685 U4UAM5 A0A336MWI6 A0A336MU49 A0A2J7REX4 A0A2R7WCI1
EC Number
3.1.3.48
Pubmed
22118469
17510324
12364791
25244985
30249741
20920257
+ More
23761445 26483478 20798317 24495485 21282665 20966253 18362917 19820115 26823975 25830018 27129103 1657401 1657402 10731132 12537572 21347285 20075255 26108605 17994087 25315136 25474469 21719571 15632085 17550304 28648823 24508170 23537049
23761445 26483478 20798317 24495485 21282665 20966253 18362917 19820115 26823975 25830018 27129103 1657401 1657402 10731132 12537572 21347285 20075255 26108605 17994087 25315136 25474469 21719571 15632085 17550304 28648823 24508170 23537049
EMBL
AGBW02012175
OWR45519.1
CH477837
EAT35785.1
AAAB01008880
GFDL01009042
+ More
JAV26003.1 GFDL01009049 JAV25996.1 EAA08669.6 APCN01005052 GGFM01006595 MBW27346.1 GGFK01001238 MBW34559.1 GANO01001941 JAB57930.1 GGFL01002740 MBW66918.1 GGFL01002759 MBW66937.1 EGK97027.1 GGFJ01000390 MBW49531.1 GGFJ01000311 MBW49452.1 DS231918 EDS26250.1 AXCN02001565 QOIP01000010 RLU17864.1 ADMH02001135 ETN63979.1 AXCM01006824 JXUM01019910 JXUM01019911 JXUM01019912 KQ560558 KXJ81897.1 ODYU01008831 SOQ52776.1 GGFL01002913 MBW67091.1 GL439483 EFN67223.1 KA646689 AFP61318.1 GAMC01018227 JAB88328.1 GDHF01028645 JAI23669.1 OUUW01000011 SPP86358.1 GL762758 EFZ20845.1 KQ971338 EFA02044.2 GDHC01009939 JAQ08690.1 GBXI01011546 JAD02746.1 GDKW01000965 JAI55630.1 CCAG010010328 M80465 M80538 AE014298 BT004474 BT126065 ADTU01010120 ADTU01010121 ADTU01010122 JXJN01000507 JXJN01000508 JRES01001303 KNC23771.1 CH902630 EDV38451.2 GBBI01003536 JAC15176.1 CH964239 EDW82297.2 GL888235 EGI64344.1 KQ760548 OAD59929.1 GFDF01006802 JAV07282.1 CH379063 EAL32708.3 GFDF01006806 JAV07278.1 GDHF01022251 JAI30063.1 KYB27808.1 CM000162 EDX02501.2 GBYB01009662 JAG79429.1 GBXI01009525 JAD04767.1 NNAY01000335 OXU29119.1 GDHF01032239 JAI20075.1 CH916370 EDV99959.1 CH954180 EDV46021.2 AJVK01006392 CH480851 EDW51417.1 CVRI01000004 CRK87652.1 CRK87650.1 GBYB01009664 JAG79431.1 CRK87651.1 KK107167 EZA56383.1 CH933812 EDW05881.2 KB632168 ERL89353.1 UFQT01002832 SSX34115.1 SSX34114.1 NEVH01004429 PNF39381.1 KK854622 PTY17382.1
JAV26003.1 GFDL01009049 JAV25996.1 EAA08669.6 APCN01005052 GGFM01006595 MBW27346.1 GGFK01001238 MBW34559.1 GANO01001941 JAB57930.1 GGFL01002740 MBW66918.1 GGFL01002759 MBW66937.1 EGK97027.1 GGFJ01000390 MBW49531.1 GGFJ01000311 MBW49452.1 DS231918 EDS26250.1 AXCN02001565 QOIP01000010 RLU17864.1 ADMH02001135 ETN63979.1 AXCM01006824 JXUM01019910 JXUM01019911 JXUM01019912 KQ560558 KXJ81897.1 ODYU01008831 SOQ52776.1 GGFL01002913 MBW67091.1 GL439483 EFN67223.1 KA646689 AFP61318.1 GAMC01018227 JAB88328.1 GDHF01028645 JAI23669.1 OUUW01000011 SPP86358.1 GL762758 EFZ20845.1 KQ971338 EFA02044.2 GDHC01009939 JAQ08690.1 GBXI01011546 JAD02746.1 GDKW01000965 JAI55630.1 CCAG010010328 M80465 M80538 AE014298 BT004474 BT126065 ADTU01010120 ADTU01010121 ADTU01010122 JXJN01000507 JXJN01000508 JRES01001303 KNC23771.1 CH902630 EDV38451.2 GBBI01003536 JAC15176.1 CH964239 EDW82297.2 GL888235 EGI64344.1 KQ760548 OAD59929.1 GFDF01006802 JAV07282.1 CH379063 EAL32708.3 GFDF01006806 JAV07278.1 GDHF01022251 JAI30063.1 KYB27808.1 CM000162 EDX02501.2 GBYB01009662 JAG79429.1 GBXI01009525 JAD04767.1 NNAY01000335 OXU29119.1 GDHF01032239 JAI20075.1 CH916370 EDV99959.1 CH954180 EDV46021.2 AJVK01006392 CH480851 EDW51417.1 CVRI01000004 CRK87652.1 CRK87650.1 GBYB01009664 JAG79431.1 CRK87651.1 KK107167 EZA56383.1 CH933812 EDW05881.2 KB632168 ERL89353.1 UFQT01002832 SSX34115.1 SSX34114.1 NEVH01004429 PNF39381.1 KK854622 PTY17382.1
Proteomes
UP000007151
UP000008820
UP000007062
UP000075840
UP000076407
UP000076408
+ More
UP000075885 UP000075903 UP000075920 UP000075900 UP000002320 UP000075886 UP000279307 UP000000673 UP000075883 UP000069272 UP000069940 UP000249989 UP000075884 UP000075881 UP000000311 UP000075880 UP000268350 UP000075882 UP000007266 UP000192221 UP000092444 UP000000803 UP000005205 UP000092460 UP000002358 UP000037069 UP000091820 UP000007801 UP000095301 UP000007798 UP000007755 UP000001819 UP000092445 UP000095300 UP000002282 UP000215335 UP000078200 UP000001070 UP000008711 UP000092462 UP000001292 UP000183832 UP000053097 UP000009192 UP000030742 UP000235965
UP000075885 UP000075903 UP000075920 UP000075900 UP000002320 UP000075886 UP000279307 UP000000673 UP000075883 UP000069272 UP000069940 UP000249989 UP000075884 UP000075881 UP000000311 UP000075880 UP000268350 UP000075882 UP000007266 UP000192221 UP000092444 UP000000803 UP000005205 UP000092460 UP000002358 UP000037069 UP000091820 UP000007801 UP000095301 UP000007798 UP000007755 UP000001819 UP000092445 UP000095300 UP000002282 UP000215335 UP000078200 UP000001070 UP000008711 UP000092462 UP000001292 UP000183832 UP000053097 UP000009192 UP000030742 UP000235965
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A212EVL1
A0A1S4FVE7
Q16N57
A0A2C9GXN6
A0A1Q3FEP0
A0A1Q3FEM2
+ More
Q7PQL0 A0A2C9GPF3 A0A2C9H7N3 A0A2M3ZFK9 A0A2M4A1D2 W4VRK2 A0A182Y5C9 A0A2M4CPV4 A0A1S4GKI9 A0A1S4GL07 A0A2M4CNN4 A0A182PD62 A0A182V608 F5HL66 A0A2M4B965 A0A2M4B8X5 A0A2C9H7M9 A0A2C9GPF4 A0A182XG36 A0A2C9GPF8 A0A182W196 A0A182RX49 B0WFI2 A0A182QZU5 A0A3L8DBY3 W5JMK0 A0A182MFC8 A0A182FPT1 A0A182GX55 A0A182NCZ2 A0A182K559 A0A2H1WIJ0 A0A2M4CPH8 E2AHA6 A0A182ISY5 T1PDC5 W8AUE6 A0A0K8UAK8 A0A3B0KR32 E9IF67 A0A182L725 D2A2C1 A0A146LQN5 A0A0A1WVW3 A0A0P4VYC9 A0A1W4VJU9 A0A1B0G7C0 P35992-2 A0A158NAA8 A0A1B0AMN8 K7IYR7 A0A0L0BUR2 A0A1A9X001 B3MXI6 A0A1I8NIH2 A0A023F268 B4NBT8 F4WN75 A0A310STH1 A0A1L8DLG1 Q29IE5 A0A1L8DLD8 A0A1B0AHB4 A0A0K8UU60 A0A1W4VJ84 A0A139WIH4 A0A1I8P7V3 B4Q1S6 A0A0C9R9Q6 A0A0A1X0Z4 P35992 A0A1W4VKM0 A0A232FFA9 A0A0K8U159 P35992-1 A0A1A9UY21 B4JK57 B3NUA6 A0A1B0DK97 B4IK45 A0A1J1HJR5 A0A1J1HJB1 A0A1W4WAG1 A0A1W4V7G9 A0A0C9R0K3 P35992-6 A0A1J1HIC1 A0A026WK83 B4L685 U4UAM5 A0A336MWI6 A0A336MU49 A0A2J7REX4 A0A2R7WCI1
Q7PQL0 A0A2C9GPF3 A0A2C9H7N3 A0A2M3ZFK9 A0A2M4A1D2 W4VRK2 A0A182Y5C9 A0A2M4CPV4 A0A1S4GKI9 A0A1S4GL07 A0A2M4CNN4 A0A182PD62 A0A182V608 F5HL66 A0A2M4B965 A0A2M4B8X5 A0A2C9H7M9 A0A2C9GPF4 A0A182XG36 A0A2C9GPF8 A0A182W196 A0A182RX49 B0WFI2 A0A182QZU5 A0A3L8DBY3 W5JMK0 A0A182MFC8 A0A182FPT1 A0A182GX55 A0A182NCZ2 A0A182K559 A0A2H1WIJ0 A0A2M4CPH8 E2AHA6 A0A182ISY5 T1PDC5 W8AUE6 A0A0K8UAK8 A0A3B0KR32 E9IF67 A0A182L725 D2A2C1 A0A146LQN5 A0A0A1WVW3 A0A0P4VYC9 A0A1W4VJU9 A0A1B0G7C0 P35992-2 A0A158NAA8 A0A1B0AMN8 K7IYR7 A0A0L0BUR2 A0A1A9X001 B3MXI6 A0A1I8NIH2 A0A023F268 B4NBT8 F4WN75 A0A310STH1 A0A1L8DLG1 Q29IE5 A0A1L8DLD8 A0A1B0AHB4 A0A0K8UU60 A0A1W4VJ84 A0A139WIH4 A0A1I8P7V3 B4Q1S6 A0A0C9R9Q6 A0A0A1X0Z4 P35992 A0A1W4VKM0 A0A232FFA9 A0A0K8U159 P35992-1 A0A1A9UY21 B4JK57 B3NUA6 A0A1B0DK97 B4IK45 A0A1J1HJR5 A0A1J1HJB1 A0A1W4WAG1 A0A1W4V7G9 A0A0C9R0K3 P35992-6 A0A1J1HIC1 A0A026WK83 B4L685 U4UAM5 A0A336MWI6 A0A336MU49 A0A2J7REX4 A0A2R7WCI1
PDB
3S3K
E-value=4.45797e-127,
Score=1170
Ontologies
GO
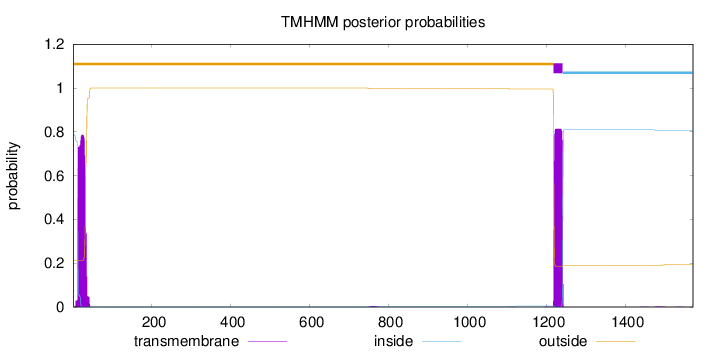
Topology
Subcellular location
Membrane
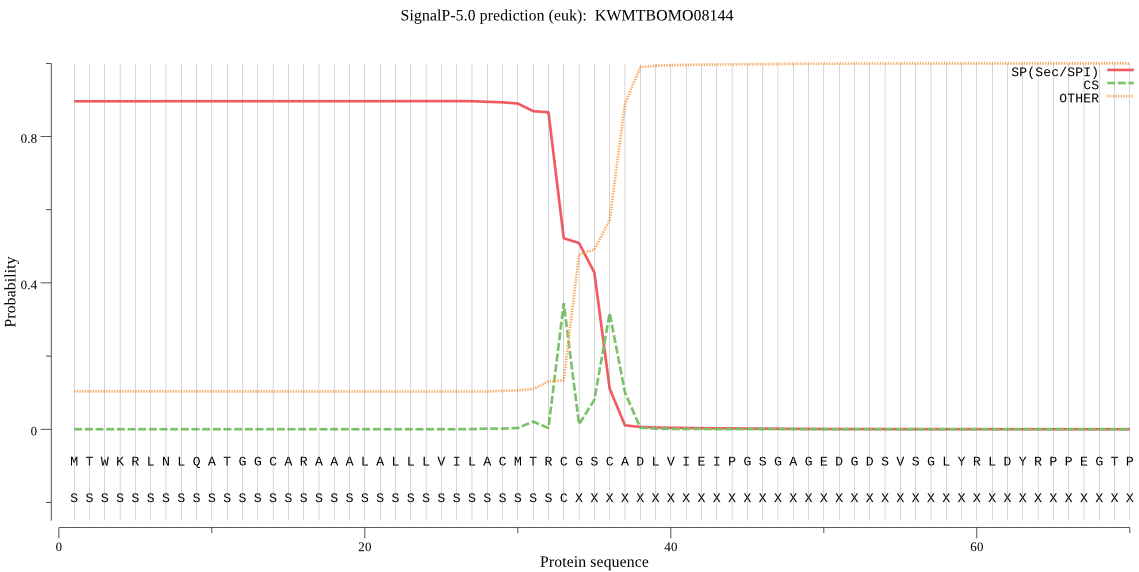
SignalP
Position: 1 - 33,
Likelihood: 0.897590
Length:
1572
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.11395
Exp number, first 60 AAs:
16.40336
Total prob of N-in:
0.78619
POSSIBLE N-term signal
sequence
outside
1 - 1218
TMhelix
1219 - 1241
inside
1242 - 1572
Population Genetic Test Statistics
Pi
0.802293
Theta
4.105012
Tajima's D
-2.184137
CLR
1623.589933
CSRT
0.00414979251037448
Interpretation
Possibly Positive selection
