Pre Gene Modal
BGIBMGA001084
Annotation
meiotic_recombination_11_[Bombyx_mori]
Full name
Double-strand break repair protein
Location in the cell
Nuclear Reliability : 2.908
Sequence
CDS
ATGATTGAGAACGACATTTCAGCGTGGTCTCCTGACGATACGCTAAGGATATTAATAGCCTCCGATATTCATCTTGGTTTCATGGAAAATGATCCGGTACGCGGAGAAGATAGCTTTATAGCTTTTGAAGAAGTTTTGTCTCTTGCGGTACAATGTGATGTTGATTTGATACTTTTAGGAGGTGATCTGTTTGATCAAGCTAAACCTTCTGTCAATTGTATGTTTAAATGTACAGAAATAATCCGCAAATATTGTCTTGGGGATAAACCGGTTAGTATAGAGTTGCTGTCTGACCAAATAAAAAATTTTTCAAGAACAGTTAACTATGAGGACCCCAATTTGAATATTTCTTACCCTATTCTCTCAATACACGGCAATCATGATGATCCCGTTGGCCAGGGCAGTGTGAGTTCTCTTGATATTCTCTCAATCACTGGGTTAGTTAATTATTTTGGTAAATGGACCGACTACACGCACGTAAGAATATCACCAGTACTTTTGCAAAAAGGTCTTACGAGGCTCGCCCTCTATGGACTTAGCCATTTAAAAGATCAGAGACTTTCACGTCTTTTTGCTGAAAAAAAAGTGGAAATGGAGAGACCCGATGAAACATTGGACTGGTTCAATTTATTTGTACTACATCAAAATCATGCTGACAGAGGACACAGTAACTATATTCCAGAAGGAGTGCTACCAAACTTTTTAGATCTTGTAGTGTGGGGCCATGAGCATGATAGCCATATTTGTCCAATGAAGGGAAATAAAACAGAGAAAGACAGCTTTTTTGTTGTACAACCAGGTAGTACTGTGGCAACCTCTTTAGCTGCAGGAGAAGCACTTCCTAAGCATTGTGGTTTATTAGAAATACATAAAGGAAATTTCAAGTTAACTCCTCTGCCACTGCAAACAGTAAGGCCTTTTATATTTAAAACCATAGTGCTTTCAGAGGAAAATATAGGCAGTGAGGATGTTAATGAAAATGAAAAGGTTCAAGAGTTTTTAAAAAACAGAGTTAATGAAGCTATTGATGAGGCTTCAAAGCTGAAGACTGCTGATCTGAGGCAGCCTTTACTACCACTCATCAGGTTAAGTATTTTTTATGAGCGAGAAAGTCAGAACTTTAATAGGATACGTTTTGGACAGAATTTTAATGGTTTGGTTGCAAATCCTAATGATCTATTAATAATGAAGAAAGAAAAAAAGATTAGAGAAAAAAGAGAACGTGATCCAGAAGAGGAAGGAGACATGACGGGTGTGGCTGCAGAAGCTGCTGATGTGGAGTCACTTTTACGTGCATATTATGAAGCTCAGCCGAAGGATAAGAGACTTTCTGTACTATCAGTCAGAGTCATAACTGATGCAGTTCGGGATTTTACCCTTAAGCACAATGAAGATGTCTTGCGGCGGGCATTTGATGCTCATAAACGTCGTTGCATTGCTGCACTATTGGAATCTACTGCAGAGACCGAAAAAGAAATTGCTGAACAACTAGAGGTTTGTAAGCGAGAGCTGGATGAAGCTGATGATGAGAAACTCCACACCTTAGTCGATGCAAGTGCAAGGCCATCAGCACCTGCAGCACAGCTTATAGTACCAGTTGTAAAGCTGCCCAATACTGTTGGCAAGCGCACCATTGTAGTATTGTCTAGTGATGAAGAAGAATTGTCGACACGATCTGGACGCGGCCGTACCACTCGTGCATCGCGCGGAAAGTCTCCTCGAGGAAAGAGCACCCGAGCGACTAAGGCTTCGGCACCTGCCCCCTCGCCGCCGGAACGCAGAACTCCGAGGCGTTCGGCAGCTCAGAAATCCACATCGTGGCTTCAGAATATCATGAACTCCAGTCGTCGCCGCGGTTCTAACGATATCGAAAATGACTCTGATTGA
Protein
MIENDISAWSPDDTLRILIASDIHLGFMENDPVRGEDSFIAFEEVLSLAVQCDVDLILLGGDLFDQAKPSVNCMFKCTEIIRKYCLGDKPVSIELLSDQIKNFSRTVNYEDPNLNISYPILSIHGNHDDPVGQGSVSSLDILSITGLVNYFGKWTDYTHVRISPVLLQKGLTRLALYGLSHLKDQRLSRLFAEKKVEMERPDETLDWFNLFVLHQNHADRGHSNYIPEGVLPNFLDLVVWGHEHDSHICPMKGNKTEKDSFFVVQPGSTVATSLAAGEALPKHCGLLEIHKGNFKLTPLPLQTVRPFIFKTIVLSEENIGSEDVNENEKVQEFLKNRVNEAIDEASKLKTADLRQPLLPLIRLSIFYERESQNFNRIRFGQNFNGLVANPNDLLIMKKEKKIREKRERDPEEEGDMTGVAAEAADVESLLRAYYEAQPKDKRLSVLSVRVITDAVRDFTLKHNEDVLRRAFDAHKRRCIAALLESTAETEKEIAEQLEVCKRELDEADDEKLHTLVDASARPSAPAAQLIVPVVKLPNTVGKRTIVVLSSDEEELSTRSGRGRTTRASRGKSPRGKSTRATKASAPAPSPPERRTPRRSAAQKSTSWLQNIMNSSRRRGSNDIENDSD
Summary
Description
Involved in DNA double-strand break repair (DSBR). Possesses single-strand endonuclease activity and double-strand-specific 3'-5' exonuclease activity. Also involved in meiotic DSB processing.
Cofactor
Mn(2+)
Similarity
Belongs to the MRE11/RAD32 family.
Uniprot
H9IV04
Q9GZJ8
A0A0L7KVX1
A0A3S2TD27
A0A212EVJ6
A0A194RG19
+ More
A0A194Q6T3 S4Q0C9 A0A2H1WIB5 A0A2A4JJT4 A0A2W1BJN3 A0A2Z5U614 A0A1Q3F3S7 B0WC67 Q0C777 A0A182G980 A0A1S4EUR8 Q16SG2 A0A2J7RR63 A0A0K8WKH3 A0A0K8U9D9 A0A0K8VFK0 A0A182QWK1 B4KH95 A0A182N7G6 Q7QID8 A0A1I8PL13 A0A182WI41 A0A1A9UJZ6 A0A0A1WSX9 W8C4T0 A0A0L0CLS9 A0A1B6E3M7 A0A1B6HCZ2 A0A182K4C4 A0A224X8V9 A0A1B0FHD3 B4N7B0 A0A0P4VXG1 B6UX44 A0A1A9YTB0 A0A0V0G7T3 A0A182R6N9 A0A023FAL1 A0A0M4EQK8 B4LQF8 A0A1A9WU80 A0A1I8M9Z7 A0A1Y1N0S8 A0A1W4VYM8 A0A1Y1N3V5 B6UX35 T1I572 B4JPJ1 B3MMU9 T1P9X4 B4P1G0 A0A0R1DM63 A0A1B6LSK9 B6UX46 B6UX41 B6UX36 B6UX48 B4G6M8 B4HX38 B6UX39 B6UX38 B4QAD4 Q29M26 B6UX47 A0A182PIW7 B6UX57 B6UX56 B6UX66 B6UX53 B3N4J0 A0A1S3HKK7 Q9XYZ4 B6UX37 B6UX42 B6UX63 B6UX40 A0A1B0BIS2 A0A182MQD9 A0A3R7LYQ0 A0A3B0JAY6 A0A336LS54 B6UX45 A0A210QGS1
A0A194Q6T3 S4Q0C9 A0A2H1WIB5 A0A2A4JJT4 A0A2W1BJN3 A0A2Z5U614 A0A1Q3F3S7 B0WC67 Q0C777 A0A182G980 A0A1S4EUR8 Q16SG2 A0A2J7RR63 A0A0K8WKH3 A0A0K8U9D9 A0A0K8VFK0 A0A182QWK1 B4KH95 A0A182N7G6 Q7QID8 A0A1I8PL13 A0A182WI41 A0A1A9UJZ6 A0A0A1WSX9 W8C4T0 A0A0L0CLS9 A0A1B6E3M7 A0A1B6HCZ2 A0A182K4C4 A0A224X8V9 A0A1B0FHD3 B4N7B0 A0A0P4VXG1 B6UX44 A0A1A9YTB0 A0A0V0G7T3 A0A182R6N9 A0A023FAL1 A0A0M4EQK8 B4LQF8 A0A1A9WU80 A0A1I8M9Z7 A0A1Y1N0S8 A0A1W4VYM8 A0A1Y1N3V5 B6UX35 T1I572 B4JPJ1 B3MMU9 T1P9X4 B4P1G0 A0A0R1DM63 A0A1B6LSK9 B6UX46 B6UX41 B6UX36 B6UX48 B4G6M8 B4HX38 B6UX39 B6UX38 B4QAD4 Q29M26 B6UX47 A0A182PIW7 B6UX57 B6UX56 B6UX66 B6UX53 B3N4J0 A0A1S3HKK7 Q9XYZ4 B6UX37 B6UX42 B6UX63 B6UX40 A0A1B0BIS2 A0A182MQD9 A0A3R7LYQ0 A0A3B0JAY6 A0A336LS54 B6UX45 A0A210QGS1
Pubmed
19121390
26227816
22118469
26354079
23622113
28756777
+ More
26760975 17510324 26483478 17519023 17994087 12364791 14747013 17210077 25830018 24495485 26108605 27129103 18984573 25474469 18057021 25315136 28004739 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685
26760975 17510324 26483478 17519023 17994087 12364791 14747013 17210077 25830018 24495485 26108605 27129103 18984573 25474469 18057021 25315136 28004739 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685
EMBL
BABH01000030
AB038379
BAB11924.1
JTDY01005187
KOB67275.1
RSAL01000324
+ More
RVE42534.1 AGBW02012175 OWR45520.1 KQ460297 KPJ16255.1 KQ459439 KPJ01084.1 GAIX01000008 JAA92552.1 ODYU01008831 SOQ52777.1 NWSH01001282 PCG71854.1 KZ150011 PZC75078.1 FX985791 BBA93678.1 GFDL01012829 JAV22216.1 DS231886 EDS43312.1 CH477186 EAT48950.1 JXUM01153566 KQ571741 KXJ68122.1 EF173378 CH477673 ABM68620.1 EAT37438.1 NEVH01000611 PNF43324.1 GDHF01000959 JAI51355.1 GDHF01029339 JAI22975.1 GDHF01014686 JAI37628.1 AXCN02000573 CH933807 EDW13312.1 AAAB01008807 EAA04178.4 GBXI01012764 JAD01528.1 GAMC01005299 JAC01257.1 JRES01000310 KNC32384.1 GEDC01004799 JAS32499.1 GECU01035307 GECU01035146 GECU01027789 GECU01025812 GECU01021565 GECU01015021 GECU01009676 JAS72399.1 JAS72560.1 JAS79917.1 JAS81894.1 JAS86141.1 JAS92685.1 JAS98030.1 GFTR01007629 JAW08797.1 CCAG010023021 CH964182 EDW80251.1 GDKW01001239 JAI55356.1 FJ219092 ACI97143.1 GECL01001929 JAP04195.1 GBBI01000171 JAC18541.1 CP012523 ALC39312.1 CH940649 EDW63408.1 KRF81091.1 GEZM01016952 JAV91018.1 GEZM01016953 JAV91016.1 FJ219083 ACI97134.1 ACPB03011506 CH916372 EDV98821.1 CH902620 EDV30974.1 KA644748 AFP59377.1 CM000157 EDW88067.2 KRJ97565.1 GEBQ01013306 JAT26671.1 FJ219094 FJ219098 FJ219100 FJ219102 FJ219103 FJ219106 FJ219109 FJ219110 FJ219113 ACI97145.1 ACI97149.1 ACI97151.1 ACI97153.1 ACI97154.1 ACI97157.1 ACI97160.1 ACI97161.1 ACI97164.1 FJ219089 ACI97140.1 FJ219084 ACI97135.1 FJ219096 FJ219097 FJ219108 FJ219115 ACI97147.1 ACI97148.1 ACI97159.1 ACI97166.1 CH479180 EDW29142.1 CH480818 EDW52583.1 FJ219087 ACI97138.1 FJ219086 ACI97137.1 CM000361 CM002910 EDX04689.1 KMY89749.1 CH379060 EAL33869.2 FJ219095 ACI97146.1 FJ219105 ACI97156.1 FJ219104 ACI97155.1 FJ219114 ACI97165.1 FJ219101 ACI97152.1 CH954177 EDV58902.1 AF132144 AE014134 AAD33591.1 AAF53093.1 FJ219085 ACI97136.1 FJ219090 ACI97141.1 FJ219111 ACI97162.1 FJ219088 ACI97139.1 JXJN01015151 JXJN01015152 JXJN01015153 JXJN01015154 AXCM01002970 QCYY01002879 ROT66880.1 OUUW01000004 SPP79507.1 UFQT01000144 SSX20836.1 FJ219093 ACI97144.1 NEDP02003741 OWF47973.1
RVE42534.1 AGBW02012175 OWR45520.1 KQ460297 KPJ16255.1 KQ459439 KPJ01084.1 GAIX01000008 JAA92552.1 ODYU01008831 SOQ52777.1 NWSH01001282 PCG71854.1 KZ150011 PZC75078.1 FX985791 BBA93678.1 GFDL01012829 JAV22216.1 DS231886 EDS43312.1 CH477186 EAT48950.1 JXUM01153566 KQ571741 KXJ68122.1 EF173378 CH477673 ABM68620.1 EAT37438.1 NEVH01000611 PNF43324.1 GDHF01000959 JAI51355.1 GDHF01029339 JAI22975.1 GDHF01014686 JAI37628.1 AXCN02000573 CH933807 EDW13312.1 AAAB01008807 EAA04178.4 GBXI01012764 JAD01528.1 GAMC01005299 JAC01257.1 JRES01000310 KNC32384.1 GEDC01004799 JAS32499.1 GECU01035307 GECU01035146 GECU01027789 GECU01025812 GECU01021565 GECU01015021 GECU01009676 JAS72399.1 JAS72560.1 JAS79917.1 JAS81894.1 JAS86141.1 JAS92685.1 JAS98030.1 GFTR01007629 JAW08797.1 CCAG010023021 CH964182 EDW80251.1 GDKW01001239 JAI55356.1 FJ219092 ACI97143.1 GECL01001929 JAP04195.1 GBBI01000171 JAC18541.1 CP012523 ALC39312.1 CH940649 EDW63408.1 KRF81091.1 GEZM01016952 JAV91018.1 GEZM01016953 JAV91016.1 FJ219083 ACI97134.1 ACPB03011506 CH916372 EDV98821.1 CH902620 EDV30974.1 KA644748 AFP59377.1 CM000157 EDW88067.2 KRJ97565.1 GEBQ01013306 JAT26671.1 FJ219094 FJ219098 FJ219100 FJ219102 FJ219103 FJ219106 FJ219109 FJ219110 FJ219113 ACI97145.1 ACI97149.1 ACI97151.1 ACI97153.1 ACI97154.1 ACI97157.1 ACI97160.1 ACI97161.1 ACI97164.1 FJ219089 ACI97140.1 FJ219084 ACI97135.1 FJ219096 FJ219097 FJ219108 FJ219115 ACI97147.1 ACI97148.1 ACI97159.1 ACI97166.1 CH479180 EDW29142.1 CH480818 EDW52583.1 FJ219087 ACI97138.1 FJ219086 ACI97137.1 CM000361 CM002910 EDX04689.1 KMY89749.1 CH379060 EAL33869.2 FJ219095 ACI97146.1 FJ219105 ACI97156.1 FJ219104 ACI97155.1 FJ219114 ACI97165.1 FJ219101 ACI97152.1 CH954177 EDV58902.1 AF132144 AE014134 AAD33591.1 AAF53093.1 FJ219085 ACI97136.1 FJ219090 ACI97141.1 FJ219111 ACI97162.1 FJ219088 ACI97139.1 JXJN01015151 JXJN01015152 JXJN01015153 JXJN01015154 AXCM01002970 QCYY01002879 ROT66880.1 OUUW01000004 SPP79507.1 UFQT01000144 SSX20836.1 FJ219093 ACI97144.1 NEDP02003741 OWF47973.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000218220 UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000075886 UP000009192 UP000075884 UP000007062 UP000095300 UP000075920 UP000078200 UP000037069 UP000075881 UP000092444 UP000007798 UP000092443 UP000075900 UP000092553 UP000008792 UP000091820 UP000095301 UP000192221 UP000015103 UP000001070 UP000007801 UP000002282 UP000008744 UP000001292 UP000000304 UP000001819 UP000075885 UP000008711 UP000085678 UP000000803 UP000092460 UP000075883 UP000283509 UP000268350 UP000242188
UP000218220 UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000075886 UP000009192 UP000075884 UP000007062 UP000095300 UP000075920 UP000078200 UP000037069 UP000075881 UP000092444 UP000007798 UP000092443 UP000075900 UP000092553 UP000008792 UP000091820 UP000095301 UP000192221 UP000015103 UP000001070 UP000007801 UP000002282 UP000008744 UP000001292 UP000000304 UP000001819 UP000075885 UP000008711 UP000085678 UP000000803 UP000092460 UP000075883 UP000283509 UP000268350 UP000242188
Interpro
Gene 3D
ProteinModelPortal
H9IV04
Q9GZJ8
A0A0L7KVX1
A0A3S2TD27
A0A212EVJ6
A0A194RG19
+ More
A0A194Q6T3 S4Q0C9 A0A2H1WIB5 A0A2A4JJT4 A0A2W1BJN3 A0A2Z5U614 A0A1Q3F3S7 B0WC67 Q0C777 A0A182G980 A0A1S4EUR8 Q16SG2 A0A2J7RR63 A0A0K8WKH3 A0A0K8U9D9 A0A0K8VFK0 A0A182QWK1 B4KH95 A0A182N7G6 Q7QID8 A0A1I8PL13 A0A182WI41 A0A1A9UJZ6 A0A0A1WSX9 W8C4T0 A0A0L0CLS9 A0A1B6E3M7 A0A1B6HCZ2 A0A182K4C4 A0A224X8V9 A0A1B0FHD3 B4N7B0 A0A0P4VXG1 B6UX44 A0A1A9YTB0 A0A0V0G7T3 A0A182R6N9 A0A023FAL1 A0A0M4EQK8 B4LQF8 A0A1A9WU80 A0A1I8M9Z7 A0A1Y1N0S8 A0A1W4VYM8 A0A1Y1N3V5 B6UX35 T1I572 B4JPJ1 B3MMU9 T1P9X4 B4P1G0 A0A0R1DM63 A0A1B6LSK9 B6UX46 B6UX41 B6UX36 B6UX48 B4G6M8 B4HX38 B6UX39 B6UX38 B4QAD4 Q29M26 B6UX47 A0A182PIW7 B6UX57 B6UX56 B6UX66 B6UX53 B3N4J0 A0A1S3HKK7 Q9XYZ4 B6UX37 B6UX42 B6UX63 B6UX40 A0A1B0BIS2 A0A182MQD9 A0A3R7LYQ0 A0A3B0JAY6 A0A336LS54 B6UX45 A0A210QGS1
A0A194Q6T3 S4Q0C9 A0A2H1WIB5 A0A2A4JJT4 A0A2W1BJN3 A0A2Z5U614 A0A1Q3F3S7 B0WC67 Q0C777 A0A182G980 A0A1S4EUR8 Q16SG2 A0A2J7RR63 A0A0K8WKH3 A0A0K8U9D9 A0A0K8VFK0 A0A182QWK1 B4KH95 A0A182N7G6 Q7QID8 A0A1I8PL13 A0A182WI41 A0A1A9UJZ6 A0A0A1WSX9 W8C4T0 A0A0L0CLS9 A0A1B6E3M7 A0A1B6HCZ2 A0A182K4C4 A0A224X8V9 A0A1B0FHD3 B4N7B0 A0A0P4VXG1 B6UX44 A0A1A9YTB0 A0A0V0G7T3 A0A182R6N9 A0A023FAL1 A0A0M4EQK8 B4LQF8 A0A1A9WU80 A0A1I8M9Z7 A0A1Y1N0S8 A0A1W4VYM8 A0A1Y1N3V5 B6UX35 T1I572 B4JPJ1 B3MMU9 T1P9X4 B4P1G0 A0A0R1DM63 A0A1B6LSK9 B6UX46 B6UX41 B6UX36 B6UX48 B4G6M8 B4HX38 B6UX39 B6UX38 B4QAD4 Q29M26 B6UX47 A0A182PIW7 B6UX57 B6UX56 B6UX66 B6UX53 B3N4J0 A0A1S3HKK7 Q9XYZ4 B6UX37 B6UX42 B6UX63 B6UX40 A0A1B0BIS2 A0A182MQD9 A0A3R7LYQ0 A0A3B0JAY6 A0A336LS54 B6UX45 A0A210QGS1
PDB
3T1I
E-value=3.3774e-103,
Score=960
Ontologies
KEGG
PATHWAY
GO
Topology
Subcellular location
Nucleus
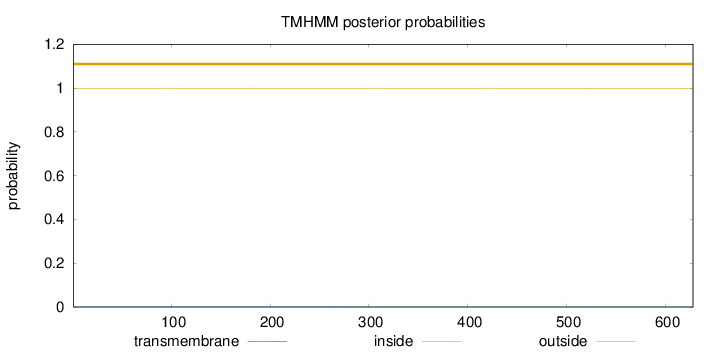
Length:
628
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00516
Exp number, first 60 AAs:
0.00044
Total prob of N-in:
0.00069
outside
1 - 628
Population Genetic Test Statistics
Pi
1.67634
Theta
4.293619
Tajima's D
-1.703047
CLR
832.811501
CSRT
0.0301484925753712
Interpretation
Possibly Positive selection
