Gene
KWMTBOMO08141 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001085
Annotation
PREDICTED:_transient_receptor_potential-gamma_protein_[Bombyx_mori]
Full name
Transient receptor potential-gamma protein
Alternative Name
Transient receptor potential cation channel gamma
Location in the cell
PlasmaMembrane Reliability : 3.028
Sequence
CDS
ATGCTGTTGCAACAGCTGAGCAAAAGCGGCGCCTCCGAGATGTCCGGCGGCAGTGGCGGCGGCGGCTCCAGTGCTGACGGGCGGCGTCCCTCCACCTGCCGCGTGGACATGGGCGCGCTGCTGGCGTCCGAGCCGCGCCCGCTCGACAGCAAGGTTAAGCGGCACAGCATCCACGGGATGACGGAAGAAGAAAATGTGGTTCGACCACATCAGGAGATGGCCGTGCTGTCTTTGGAGGAGAAGAAGTACCTGCTGGGCGTGGAGCGAGGAGACGTGGCGGGGACGCGGCGCGTGCTGCAGCGGGCCCGCGACACCGGACACATCAACGTCGACTGTGTGGACCCGCTCGGACGCAGCGCGCTGCTCATGGCCATTGACAACGAGAACCTCGAGATGGTGGAGCTGCTCCTCGAGTTCGGCGTGGAGACCCGCGACGCACTGCTGCACGCCATCTCCGAGGAGTTCGTCGAGGCCGTGGAGGCCCTGCTCGACCACGAGGAGCGGACCAGGAAGCCCGGCGAACCGAACAGTTGGGAGGCGCTTCCGCCGGAGACGGCCACGTTTACGTCTGACATCACGCCGCTGATCCTGGCGGCGCACCGGGACAGCTACGAGATCATCAAGCTGCTGCTGGACCGCGGCGCGCAGCTGCCCGAGCCGCACGACGTGCGCTGCGGCTGCGACGACTGCGTGCGCTCCCGCCGCGAGGACTCGCTGCGACACTCCCGCTCCAGGATCAACGCGTACCGCGCGCTCGCCTCCCCTTCGCTCATCGCCCTATCCTCCAAGGACCCCATTTTAACCGCTTTCGAGCTGTCGTGGGAGCTGCGCCGGTTGTCCGCATTGGAACATGAGTTCAAAACGGAATACCAGGAGCTCCGCGTTCAGTGTCAGGAGTTCGCGACTGCACTGTTGGATCACACGCGGACGTCTAATGAACTTCAGATCCTGCTGAATCACGAGAAGGGTGCTTCACCTCAGGCACCGCTCACTGAACCAGGCGCTCCCGAGCGGATGCGCTTGTCGCGGCTCAAGCTTGCCATCAAGTTGCGACAAAAGAAGTTCGTGGCGCACCCGAACGTGCAGCAGCTGCTGGCGTCGATCTGGTACGAGAGCGTGCCCGGGTTCCGCCGCAAGAACATGCTGCTGCAGGCGGCGGAGATGGTGCGGATCGGCGCCATGTTCCCGCTCTACTCGCTCGCCTACATCGCGGCGCCGCACTCCGCCGTCGGACGCACGCTCAGGAAGCCCTTCATCAAATTCCTCTGCCACTCCGCCAGTTACTTTATGTTTCTATTTCTGTTGATCCTGGCGTCGCAGCGCATCGAGACGGCGCCGGGCGGGCTGCTGTGGGACGTCTCCCACGACGAGCCGCTCTCCCGGCGCGGCTCCATGCCCTCCATAGTCGAGTGGCTCATACTCGCCTGGGTCAGCGGCTTAATCTGGAGCGAAGTGAAGCAGCTGTGGGACATGGGGCTGAGAGAGTACGTACATGACATGTGGAACGTCATCGACTTCGTCACCAACTCGCTGTACGTGGCCACCGTGGCGCTCAGGATCGTCTCGCACTACCAGGTTCGTCGCGAGATGGCGATGGGCCTGCAGTGGAACCAACCTCGCGAGAAGTGGGACGCTTGGGACCCGATGCTGCTCTCCGAGGGGCTGTTCTCGGCCGCCAACATCTTCAGCAGTCTCAAGTTGGTGTACATCTTCAGCGTGAACCCTCACCTCGGTCCGTTGCAGGTGTCGCTGAGTCGGATGGTGTTGGACATCCTCAAGTTTTTTGTCTTGGACATACTCGTCATATTTGCGTTCTCATGCGGGCTAAACCAGTTGTTGTGGTACTACGCGGACATGGAGAAGAAGCGCTGCACGGTGGGCACCGCGCTCACGCCCAACGGCACCCTGCCCGACCCGGACGCCTGTGTCGTCTGGAGACGGTTTGCTAATTTGTTCGAGACAATGCAGACGCTGTTCTGGGCTGCGTTCGGGCTGGTGGACTTGGATTCGTTCGAGCTCGACGGGATCAAGATCTTCACGCGGTTCTGGGGGATGCTCATGTTCGGCACGTACGCCGTCATCAACGTCATCGTGCTGCTCAACTTGCTCATCGCCATGATGAACCACTCCTATCAGCTGATATCCGAGCGTGCAGATGTGGAATGGAAGTTCGCCCGCAGCAAGCTGTGGATCAGCTACTTTGAGGAGGGTGGAACTGCGCCCCCGCCCTTCAACGTGCTGCCCTCGCCCAAGTCCCTGCTGTACGCCTGGCGGTGGCTGCAGCGCCGGCTCTGTGGGCACGCCCGCGCCAAGCGAGAACATATGCGGACTATTAGGAGAAAGGTGAAGCAAGCCAATGAGAGAGACTTTCGATATCAGGCTATAATGCGGAACCTGGTGCGTCGCTACGTGACGGTCCAGCAGCGGCGCGCGGAGAGCGGCGGCGTCACCGAGGACGACGTCAACGAGATCAAGCAGGACGTCTCCGCCTTCCGCTGCGAGCTCGTCGAGATCCTGCGCAACTCCGGGATGAACACCTCCACAGCCAACGCGGGAGCTCCCGGCGGCGGAGGCGGCAAAAAGAATCGTCAGAAGGAGCGACGCCTCATGAAGGGATTCAACATAGCTCCGGGCGGGTCCCTGGCCCCCGTGGACGAGTTCATGTCGCCCGTGTCCTGGCTGCAGCACGACCACGGCGTGCCGCACTACTCGCTGTCCACGCTGCTCGGGCCGCGCCTGCGAGCCTCCCAGTCCAGCCTCAGCGACGGACCCGGCGCCGGGATGAGTGCGTCGCGTCGCAAGCCGCAGCACAAGCGTCGCTGGGGCACAATCATCGACGCCGCTCGGGCGGCCCGTGTGTCTCGACTCATCGGCCGGTCGCGCTCTGAGGACTCTGTCTGCGATCACGCGCGGCAGTCGCCAAGCGGTAGCGAGCCTTCGGAGTCGGGCAGCGACTCGCAGCGCAGTCCGGAGCGGACGGGATCTCGGAGTGGACCCTTGCATCCGTTGACGGCACTGGCGGCCCTCAAGCGGAAGCGAAAGAAATTCTCGGATTCCCGGCGGCCCGAAGCCACGGTCCGGCCCGTTCCGGCGCCTGAGAGTCTGCAGCGCGCCAGTTCGGTGCCGGCTCGCGTGCCCCCCGCCGCCCCGCCCCGCGCCCCTCCGCCGCCCGCCGTGTCGCCCTCCACCACCGCCGAGAGCGTGGCCGCGCGCGGAGGCAGCCGGGAGCCGCTGCTGGCCAGCCTCGACGACGAAGCGCACAAGCGGGACGGCTGCGGCCAGTGCGGGTACGAGTCGGGGCCGCAGGGCGGCGGGGAGGCGGGGGGCGGGGCGGGCGAGGCGGAGGGCGCGTGCGGGCGGTGCGCGGCGCTGGCGCGGCTGGCGGGCGTGACGCCGCTGGCGGGCCACGCGCCGCACCACTCCGCGGGCTGGCTCTAG
Protein
MLLQQLSKSGASEMSGGSGGGGSSADGRRPSTCRVDMGALLASEPRPLDSKVKRHSIHGMTEEENVVRPHQEMAVLSLEEKKYLLGVERGDVAGTRRVLQRARDTGHINVDCVDPLGRSALLMAIDNENLEMVELLLEFGVETRDALLHAISEEFVEAVEALLDHEERTRKPGEPNSWEALPPETATFTSDITPLILAAHRDSYEIIKLLLDRGAQLPEPHDVRCGCDDCVRSRREDSLRHSRSRINAYRALASPSLIALSSKDPILTAFELSWELRRLSALEHEFKTEYQELRVQCQEFATALLDHTRTSNELQILLNHEKGASPQAPLTEPGAPERMRLSRLKLAIKLRQKKFVAHPNVQQLLASIWYESVPGFRRKNMLLQAAEMVRIGAMFPLYSLAYIAAPHSAVGRTLRKPFIKFLCHSASYFMFLFLLILASQRIETAPGGLLWDVSHDEPLSRRGSMPSIVEWLILAWVSGLIWSEVKQLWDMGLREYVHDMWNVIDFVTNSLYVATVALRIVSHYQVRREMAMGLQWNQPREKWDAWDPMLLSEGLFSAANIFSSLKLVYIFSVNPHLGPLQVSLSRMVLDILKFFVLDILVIFAFSCGLNQLLWYYADMEKKRCTVGTALTPNGTLPDPDACVVWRRFANLFETMQTLFWAAFGLVDLDSFELDGIKIFTRFWGMLMFGTYAVINVIVLLNLLIAMMNHSYQLISERADVEWKFARSKLWISYFEEGGTAPPPFNVLPSPKSLLYAWRWLQRRLCGHARAKREHMRTIRRKVKQANERDFRYQAIMRNLVRRYVTVQQRRAESGGVTEDDVNEIKQDVSAFRCELVEILRNSGMNTSTANAGAPGGGGGKKNRQKERRLMKGFNIAPGGSLAPVDEFMSPVSWLQHDHGVPHYSLSTLLGPRLRASQSSLSDGPGAGMSASRRKPQHKRRWGTIIDAARAARVSRLIGRSRSEDSVCDHARQSPSGSEPSESGSDSQRSPERTGSRSGPLHPLTALAALKRKRKKFSDSRRPEATVRPVPAPESLQRASSVPARVPPAAPPRAPPPPAVSPSTTAESVAARGGSREPLLASLDDEAHKRDGCGQCGYESGPQGGGEAGGGAGEAEGACGRCAALARLAGVTPLAGHAPHHSAGWL
Summary
Description
A light-sensitive calcium channel that is required for inositide-mediated Ca(2+) entry in the retina during phospholipase C (PLC)-mediated phototransduction (By similarity). Forms a regulated cation channel when heteromultimerized with trpl.
Subunit
Interacts preferentially with trpl and interacts to a lower extent with trp.
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Belongs to the transient receptor (TC 1.A.4) family. STrpC subfamily.
Belongs to the transient receptor (TC 1.A.4) family. STrpC subfamily.
Keywords
ANK repeat
Calcium
Calcium channel
Calcium transport
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Repeat
Sensory transduction
Transmembrane
Transmembrane helix
Transport
Vision
Feature
chain Transient receptor potential-gamma protein
Uniprot
H9IV05
A0A212EVK1
A0A2H1WAA9
A0A2A4J9Y4
A0A2W1BXK2
A0A182RRP8
+ More
A0A182WHD9 A0A1S4F9E4 A0A182TJL8 A0A1S4GYA0 A0A182L0E3 A0A182PAQ8 A0A182X110 A0A195F2N6 A0A182IC15 A0A182USM7 A0A182NES6 A0A195BR92 A0A0L7KX81 A0A158P1R4 A0A3L8D875 A0A182QJA4 A0A182Y9Z8 E2BGT4 Q17B94 A0A195DC69 A0A182F5L0 A0A088AV64 A0A2M3YXM0 A0A2M4CSL6 A0A067RHQ7 Q7Q4K2 A0A182K9S5 A0A2A3ER74 A0A2M3YXM2 A0A2M4CSW9 A0A2M4CSG3 B7YZW4 A0A2M3ZZU4 A0A2M4BA84 D0QWH5 B4G7Q9 A0A3B0JBF9 A0A034WJC1 U5ENB5 A0A1I8NXJ4 A0A1I8MKX1 A0A0M4EDQ7 A0A384TJQ6 A0A1B0BV64 A0A0L0C5Y3 A0A0R1DV77 B4P9C4 A0A0J9TPI4 A0A0J9R2R7 B3MKM4 Q9VJJ7 A0A1W4VJZ0 A0A1W4VK61 A0A0Q5VXN4 B3NNI2 B4KKT8 A0A0P8XX94 Q29LQ7 A0A0R3NTR5 B4LVK6 A0A1W4VY66 B4NLT7 T1H8K7 A0A0L7RKN9 A0A0M8ZWD1 B4JZ78 A0A0C9R3K2 Q5YJT9 A0A151WQR9 A0A0C9QGE7 B3G505 K7IZ88 E0VCF7 A0A0J7NYX8 A0A026W1F3 E2AZK2 A0A2J7PZ76 A0A1B0GFN2 A0A154NZ64 A0A226E7F5 E9GZL1 A0A0P6HFY0 A0A164TG31 A0A0P6AAK5 A0A0P6GJB5 A0A0P5DNR9 A0A0P5ZY57 A0A0N7ZKT9 A0A059XPJ5 A0A1V9Y157 B7PBX3 A0A336MW87 A0A336M9W2
A0A182WHD9 A0A1S4F9E4 A0A182TJL8 A0A1S4GYA0 A0A182L0E3 A0A182PAQ8 A0A182X110 A0A195F2N6 A0A182IC15 A0A182USM7 A0A182NES6 A0A195BR92 A0A0L7KX81 A0A158P1R4 A0A3L8D875 A0A182QJA4 A0A182Y9Z8 E2BGT4 Q17B94 A0A195DC69 A0A182F5L0 A0A088AV64 A0A2M3YXM0 A0A2M4CSL6 A0A067RHQ7 Q7Q4K2 A0A182K9S5 A0A2A3ER74 A0A2M3YXM2 A0A2M4CSW9 A0A2M4CSG3 B7YZW4 A0A2M3ZZU4 A0A2M4BA84 D0QWH5 B4G7Q9 A0A3B0JBF9 A0A034WJC1 U5ENB5 A0A1I8NXJ4 A0A1I8MKX1 A0A0M4EDQ7 A0A384TJQ6 A0A1B0BV64 A0A0L0C5Y3 A0A0R1DV77 B4P9C4 A0A0J9TPI4 A0A0J9R2R7 B3MKM4 Q9VJJ7 A0A1W4VJZ0 A0A1W4VK61 A0A0Q5VXN4 B3NNI2 B4KKT8 A0A0P8XX94 Q29LQ7 A0A0R3NTR5 B4LVK6 A0A1W4VY66 B4NLT7 T1H8K7 A0A0L7RKN9 A0A0M8ZWD1 B4JZ78 A0A0C9R3K2 Q5YJT9 A0A151WQR9 A0A0C9QGE7 B3G505 K7IZ88 E0VCF7 A0A0J7NYX8 A0A026W1F3 E2AZK2 A0A2J7PZ76 A0A1B0GFN2 A0A154NZ64 A0A226E7F5 E9GZL1 A0A0P6HFY0 A0A164TG31 A0A0P6AAK5 A0A0P6GJB5 A0A0P5DNR9 A0A0P5ZY57 A0A0N7ZKT9 A0A059XPJ5 A0A1V9Y157 B7PBX3 A0A336MW87 A0A336M9W2
Pubmed
19121390
22118469
28756777
12364791
20966253
26227816
+ More
21347285 30249741 25244985 20798317 17510324 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19859648 17994087 25348373 25315136 26108605 17550304 22936249 18057021 10896160 15632085 16319060 19191929 20075255 20566863 24508170 21292972 28327890
21347285 30249741 25244985 20798317 17510324 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19859648 17994087 25348373 25315136 26108605 17550304 22936249 18057021 10896160 15632085 16319060 19191929 20075255 20566863 24508170 21292972 28327890
EMBL
BABH01000036
BABH01000037
BABH01000038
AGBW02012175
OWR45522.1
ODYU01007315
+ More
SOQ49990.1 NWSH01002346 PCG68478.1 KZ149891 PZC79051.1 AAAB01008964 KQ981855 KYN34835.1 APCN01000771 KQ976418 KYM89481.1 JTDY01004876 KOB67644.1 ADTU01001096 ADTU01001097 ADTU01001098 ADTU01001099 ADTU01001100 ADTU01001101 ADTU01001102 ADTU01001103 ADTU01001104 ADTU01001105 QOIP01000012 RLU16302.1 AXCN02000840 GL448228 EFN85066.1 CH477324 EAT43552.1 KQ980989 KYN10488.1 GGFM01000266 MBW21017.1 GGFL01004091 MBW68269.1 KK852631 KDR19850.1 EAA12444.4 KZ288192 PBC34293.1 GGFM01000262 MBW21013.1 GGFL01004093 MBW68271.1 GGFL01004092 MBW68270.1 AE014134 ACL83036.2 GGFK01000746 MBW34067.1 GGFJ01000823 MBW49964.1 FJ821029 ACN94674.1 CH479180 EDW29330.1 OUUW01000004 SPP79365.1 GAKP01004193 JAC54759.1 GANO01000663 JAB59208.1 CP012523 ALC40022.1 KX249690 AOR81467.1 JXJN01021148 JRES01000842 KNC27808.1 CM000158 KRJ99073.1 EDW90253.2 KRJ99074.1 CM002910 KMY90538.1 KMY90540.1 KMY90537.1 KMY90539.1 CH902620 EDV32537.2 KPU74022.1 KPU74023.1 AJ277967 AJ277968 CH954179 KQS62953.1 EDV56633.2 KQS62954.1 CH933807 EDW11668.2 KPU74021.1 CH379060 EAL33988.3 KRT04378.1 CH940649 EDW64400.2 CH964274 EDW85326.2 ACPB03022385 KQ414572 KOC71316.1 KQ435850 KOX70909.1 CH916379 EDV94187.1 GBYB01002525 JAG72292.1 AY387857 AAR26327.1 KQ982819 KYQ50204.1 GBYB01002524 JAG72291.1 EU649776 ACD69414.1 AAZO01001036 DS235053 EEB11063.1 LBMM01000754 KMQ97565.1 KK107488 EZA49892.1 GL444248 EFN61135.1 NEVH01020337 PNF21643.1 CCAG010018206 KQ434778 KZC04354.1 LNIX01000006 OXA52821.1 GL732578 EFX75016.1 GDIQ01019517 JAN75220.1 LRGB01001801 KZS10426.1 GDIP01033238 JAM70477.1 GDIQ01042893 JAN51844.1 GDIP01153419 JAJ69983.1 GDIQ01156296 GDIP01038044 JAK95429.1 JAM65671.1 GDIP01235625 JAI87776.1 KJ572528 AIA22827.1 MNPL01001084 OQR79469.1 ABJB010218724 ABJB010332204 ABJB010546725 ABJB011014079 DS679906 EEC04095.1 UFQT01002971 SSX34380.1 UFQT01000733 SSX26810.1
SOQ49990.1 NWSH01002346 PCG68478.1 KZ149891 PZC79051.1 AAAB01008964 KQ981855 KYN34835.1 APCN01000771 KQ976418 KYM89481.1 JTDY01004876 KOB67644.1 ADTU01001096 ADTU01001097 ADTU01001098 ADTU01001099 ADTU01001100 ADTU01001101 ADTU01001102 ADTU01001103 ADTU01001104 ADTU01001105 QOIP01000012 RLU16302.1 AXCN02000840 GL448228 EFN85066.1 CH477324 EAT43552.1 KQ980989 KYN10488.1 GGFM01000266 MBW21017.1 GGFL01004091 MBW68269.1 KK852631 KDR19850.1 EAA12444.4 KZ288192 PBC34293.1 GGFM01000262 MBW21013.1 GGFL01004093 MBW68271.1 GGFL01004092 MBW68270.1 AE014134 ACL83036.2 GGFK01000746 MBW34067.1 GGFJ01000823 MBW49964.1 FJ821029 ACN94674.1 CH479180 EDW29330.1 OUUW01000004 SPP79365.1 GAKP01004193 JAC54759.1 GANO01000663 JAB59208.1 CP012523 ALC40022.1 KX249690 AOR81467.1 JXJN01021148 JRES01000842 KNC27808.1 CM000158 KRJ99073.1 EDW90253.2 KRJ99074.1 CM002910 KMY90538.1 KMY90540.1 KMY90537.1 KMY90539.1 CH902620 EDV32537.2 KPU74022.1 KPU74023.1 AJ277967 AJ277968 CH954179 KQS62953.1 EDV56633.2 KQS62954.1 CH933807 EDW11668.2 KPU74021.1 CH379060 EAL33988.3 KRT04378.1 CH940649 EDW64400.2 CH964274 EDW85326.2 ACPB03022385 KQ414572 KOC71316.1 KQ435850 KOX70909.1 CH916379 EDV94187.1 GBYB01002525 JAG72292.1 AY387857 AAR26327.1 KQ982819 KYQ50204.1 GBYB01002524 JAG72291.1 EU649776 ACD69414.1 AAZO01001036 DS235053 EEB11063.1 LBMM01000754 KMQ97565.1 KK107488 EZA49892.1 GL444248 EFN61135.1 NEVH01020337 PNF21643.1 CCAG010018206 KQ434778 KZC04354.1 LNIX01000006 OXA52821.1 GL732578 EFX75016.1 GDIQ01019517 JAN75220.1 LRGB01001801 KZS10426.1 GDIP01033238 JAM70477.1 GDIQ01042893 JAN51844.1 GDIP01153419 JAJ69983.1 GDIQ01156296 GDIP01038044 JAK95429.1 JAM65671.1 GDIP01235625 JAI87776.1 KJ572528 AIA22827.1 MNPL01001084 OQR79469.1 ABJB010218724 ABJB010332204 ABJB010546725 ABJB011014079 DS679906 EEC04095.1 UFQT01002971 SSX34380.1 UFQT01000733 SSX26810.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000075900
UP000075920
UP000075902
+ More
UP000075882 UP000075885 UP000076407 UP000078541 UP000075840 UP000075903 UP000075884 UP000078540 UP000037510 UP000005205 UP000279307 UP000075886 UP000076408 UP000008237 UP000008820 UP000078492 UP000069272 UP000005203 UP000027135 UP000007062 UP000075881 UP000242457 UP000000803 UP000008744 UP000268350 UP000095300 UP000095301 UP000092553 UP000092460 UP000037069 UP000002282 UP000007801 UP000192221 UP000008711 UP000009192 UP000001819 UP000008792 UP000007798 UP000015103 UP000053825 UP000053105 UP000001070 UP000075809 UP000002358 UP000009046 UP000036403 UP000053097 UP000000311 UP000235965 UP000092444 UP000076502 UP000198287 UP000000305 UP000076858 UP000192247 UP000001555
UP000075882 UP000075885 UP000076407 UP000078541 UP000075840 UP000075903 UP000075884 UP000078540 UP000037510 UP000005205 UP000279307 UP000075886 UP000076408 UP000008237 UP000008820 UP000078492 UP000069272 UP000005203 UP000027135 UP000007062 UP000075881 UP000242457 UP000000803 UP000008744 UP000268350 UP000095300 UP000095301 UP000092553 UP000092460 UP000037069 UP000002282 UP000007801 UP000192221 UP000008711 UP000009192 UP000001819 UP000008792 UP000007798 UP000015103 UP000053825 UP000053105 UP000001070 UP000075809 UP000002358 UP000009046 UP000036403 UP000053097 UP000000311 UP000235965 UP000092444 UP000076502 UP000198287 UP000000305 UP000076858 UP000192247 UP000001555
Pfam
Interpro
IPR020683
Ankyrin_rpt-contain_dom
+ More
IPR036770 Ankyrin_rpt-contain_sf
IPR005821 Ion_trans_dom
IPR013555 TRP_dom
IPR002153 TRPC_channel
IPR002110 Ankyrin_rpt
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR036640 ABC1_TM_sf
IPR003593 AAA+_ATPase
IPR036770 Ankyrin_rpt-contain_sf
IPR005821 Ion_trans_dom
IPR013555 TRP_dom
IPR002153 TRPC_channel
IPR002110 Ankyrin_rpt
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR036640 ABC1_TM_sf
IPR003593 AAA+_ATPase
Gene 3D
CDD
ProteinModelPortal
H9IV05
A0A212EVK1
A0A2H1WAA9
A0A2A4J9Y4
A0A2W1BXK2
A0A182RRP8
+ More
A0A182WHD9 A0A1S4F9E4 A0A182TJL8 A0A1S4GYA0 A0A182L0E3 A0A182PAQ8 A0A182X110 A0A195F2N6 A0A182IC15 A0A182USM7 A0A182NES6 A0A195BR92 A0A0L7KX81 A0A158P1R4 A0A3L8D875 A0A182QJA4 A0A182Y9Z8 E2BGT4 Q17B94 A0A195DC69 A0A182F5L0 A0A088AV64 A0A2M3YXM0 A0A2M4CSL6 A0A067RHQ7 Q7Q4K2 A0A182K9S5 A0A2A3ER74 A0A2M3YXM2 A0A2M4CSW9 A0A2M4CSG3 B7YZW4 A0A2M3ZZU4 A0A2M4BA84 D0QWH5 B4G7Q9 A0A3B0JBF9 A0A034WJC1 U5ENB5 A0A1I8NXJ4 A0A1I8MKX1 A0A0M4EDQ7 A0A384TJQ6 A0A1B0BV64 A0A0L0C5Y3 A0A0R1DV77 B4P9C4 A0A0J9TPI4 A0A0J9R2R7 B3MKM4 Q9VJJ7 A0A1W4VJZ0 A0A1W4VK61 A0A0Q5VXN4 B3NNI2 B4KKT8 A0A0P8XX94 Q29LQ7 A0A0R3NTR5 B4LVK6 A0A1W4VY66 B4NLT7 T1H8K7 A0A0L7RKN9 A0A0M8ZWD1 B4JZ78 A0A0C9R3K2 Q5YJT9 A0A151WQR9 A0A0C9QGE7 B3G505 K7IZ88 E0VCF7 A0A0J7NYX8 A0A026W1F3 E2AZK2 A0A2J7PZ76 A0A1B0GFN2 A0A154NZ64 A0A226E7F5 E9GZL1 A0A0P6HFY0 A0A164TG31 A0A0P6AAK5 A0A0P6GJB5 A0A0P5DNR9 A0A0P5ZY57 A0A0N7ZKT9 A0A059XPJ5 A0A1V9Y157 B7PBX3 A0A336MW87 A0A336M9W2
A0A182WHD9 A0A1S4F9E4 A0A182TJL8 A0A1S4GYA0 A0A182L0E3 A0A182PAQ8 A0A182X110 A0A195F2N6 A0A182IC15 A0A182USM7 A0A182NES6 A0A195BR92 A0A0L7KX81 A0A158P1R4 A0A3L8D875 A0A182QJA4 A0A182Y9Z8 E2BGT4 Q17B94 A0A195DC69 A0A182F5L0 A0A088AV64 A0A2M3YXM0 A0A2M4CSL6 A0A067RHQ7 Q7Q4K2 A0A182K9S5 A0A2A3ER74 A0A2M3YXM2 A0A2M4CSW9 A0A2M4CSG3 B7YZW4 A0A2M3ZZU4 A0A2M4BA84 D0QWH5 B4G7Q9 A0A3B0JBF9 A0A034WJC1 U5ENB5 A0A1I8NXJ4 A0A1I8MKX1 A0A0M4EDQ7 A0A384TJQ6 A0A1B0BV64 A0A0L0C5Y3 A0A0R1DV77 B4P9C4 A0A0J9TPI4 A0A0J9R2R7 B3MKM4 Q9VJJ7 A0A1W4VJZ0 A0A1W4VK61 A0A0Q5VXN4 B3NNI2 B4KKT8 A0A0P8XX94 Q29LQ7 A0A0R3NTR5 B4LVK6 A0A1W4VY66 B4NLT7 T1H8K7 A0A0L7RKN9 A0A0M8ZWD1 B4JZ78 A0A0C9R3K2 Q5YJT9 A0A151WQR9 A0A0C9QGE7 B3G505 K7IZ88 E0VCF7 A0A0J7NYX8 A0A026W1F3 E2AZK2 A0A2J7PZ76 A0A1B0GFN2 A0A154NZ64 A0A226E7F5 E9GZL1 A0A0P6HFY0 A0A164TG31 A0A0P6AAK5 A0A0P6GJB5 A0A0P5DNR9 A0A0P5ZY57 A0A0N7ZKT9 A0A059XPJ5 A0A1V9Y157 B7PBX3 A0A336MW87 A0A336M9W2
PDB
5Z96
E-value=0,
Score=1787
Ontologies
GO
GO:0005262
GO:0016021
GO:0042626
GO:0005524
GO:0006811
GO:0016020
GO:0070588
GO:0051480
GO:0015279
GO:0070679
GO:0005887
GO:0006828
GO:0034703
GO:0005261
GO:0006812
GO:0007628
GO:0043025
GO:0009416
GO:0050908
GO:1990635
GO:0050884
GO:0006816
GO:0008381
GO:0016028
GO:0016740
GO:0005515
GO:0005216
GO:0015914
GO:0006412
GO:0016876
GO:0043039
GO:0055085
PANTHER
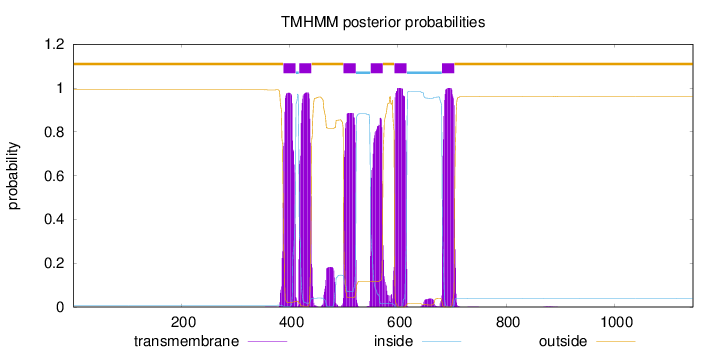
Topology
Subcellular location
Membrane
Length:
1145
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
130.89921
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00796
outside
1 - 388
TMhelix
389 - 411
inside
412 - 417
TMhelix
418 - 440
outside
441 - 499
TMhelix
500 - 522
inside
523 - 549
TMhelix
550 - 572
outside
573 - 593
TMhelix
594 - 616
inside
617 - 681
TMhelix
682 - 704
outside
705 - 1145
Population Genetic Test Statistics
Pi
1.430578
Theta
5.058776
Tajima's D
-1.949945
CLR
629.851816
CSRT
0.0138993050347483
Interpretation
Possibly Positive selection
