Gene
KWMTBOMO08140
Pre Gene Modal
BGIBMGA001076
Annotation
hypothetical_protein_KGM_13300_[Danaus_plexippus]
Full name
Caseinolytic peptidase B protein homolog
Alternative Name
Suppressor of potassium transport defect 3
Location in the cell
Cytoplasmic Reliability : 1.441 Mitochondrial Reliability : 1.526
Sequence
CDS
ATGTCGAGTCTTAGACCTGCAACGTGTTGTGTGCGTGCTTTACGACGGACGGCACGACTTGCAGGGGTGGCAGCACATCGAATATCTTCAGGATACAATAGTTCTGGAAATGAGAGGAAACAACGTTTAGATGTAAATCGTGTCACCCTAGGAGTGGGCTCATTGGCCTTGGCTCTACTCGCTGCGGATCACGTTTTTAACGAAAAAAGGTTTTTCAGAGCAGCCCAAATAGGAAATGTCGAAGAGTTAAAAAGGCAAGCGGAATACCTTATTGCTAAAGAAAGTCAGCAACCAAAGAAAGATGACGAGGGAGGCCCGGTGGATCGCCGTCACCCGCTCGGCTGGACTGCGCTCATGGTCGCGGCCGCCAACGACAAACCCGACGTCGTCCGCGAACTTATCAAACTCGGGGCCAAACCTGACCTACAGGAACAGTATGCGGGTGCGGCGACGGCCGCCTCGGCCGCCGGCATGCATCCGCTGGACGCGCTGCAACGACGGGAGGACGAGTTTTGTGGCGGCATGAACGCCCGCGCCTCCTTCCTGGGCTGGACGGCGCTGCACTACGCGGCGCTGGCCGACTCGCACGACGCCGCCGCGGCGCTGCTGGACGCCGGCGCGGACCCTGCCGCTCGCGACCACGCCGGCCGTCGCCCCCTGCACTACGCGCGGGACGCCTCGCCCACCCGCGACCTCATCCAGCGAGCCGTGCGCCGACGGGAGGAGGAAGCAGCCGCCGCCGCTGCTGAGGAGCGACGCCGCTTCCCTCTGGAGCGCCGCTTGAAGCAGTACATAGTGGGACAGCGTGCCGCCATCCACACCGTCGCCGCCGCCGTGCGCCGCAAGGAGAACGGTTGGGCCGACGACGACCATCCCCTGGTGTTCCTGTTCCTGGGGAGCTCCGGCATCGGGAAGACGGAGCTAGCGAAGCAGCTCGCCCGCTACATGCACAAGGATGACCCGGCCGCGTTCATACGTCTCGATATGTCCGAGTACCAGGAGAAACACGAGGTGGCGAAACTCATCGGAGCCCCTCCCGGATACGTGGGCCACGAGGAGGGCGGCCAGCTCACGCGGGCGCTGGCGCAGCGAGCGGACGCCGTGGTGCTGTTCGACGAGGTGGACAAGGCTCACCCCGACGTCCTCACGGTCCTGCTCCAGCTCTTTGACGAGGGCCGGCTGACGGACGGGAAGGGCAAGTTGATCGAGTGTAAGGACGCCATATTCGTGATGACGTCAAACCTGGCGGCCGACGAGATCGCGCAGTACGGGCTGCAGCTGCGCCGCGAGGCAGACGCGCTCGCCGCCACCAGGCGCGGGAAGCGGGAAGGTGAAGAAGAGGAGCAAGAGCCGACGCTAGAGGTGTCGCGGCACTTCAAGGACAACGTGGTGCGGCCCATCCTCAAGCGACAGTTCCGGCGCGACGAGTTCCTCGGTCGCATCAACGAGATCGTCTACTTCCTGCCGTTCTCTCGCGCCGAGCTGCTGGCGCTGGTGCAGGCCGAGCTGCAGCGCTGGGCGGCGCGCGCGCGCCAGCGGCACGGCATGCTGCTGCTCTGGGAGGACGACGTGCTGGGAGCCCTGGCCGACGGGTACGAGGTGCACTACGGCGCGCGCTCCATCCAGCACGAGGTGGAGCGGCGCGTGGTGAGTCAGCTGGCGCTGGCGGCGGAGCGCGGGCTGGTGGCGCGCGGCTGCACGCTGCGGCTGCGCGCGGCGCCCGCCGGCCTGGCGCTGGCGCTGCGCCCCGCCGGCGCCGCTGACTTCCAGCCGCTGCACCTGCCGCCCGCGTGA
Protein
MSSLRPATCCVRALRRTARLAGVAAHRISSGYNSSGNERKQRLDVNRVTLGVGSLALALLAADHVFNEKRFFRAAQIGNVEELKRQAEYLIAKESQQPKKDDEGGPVDRRHPLGWTALMVAAANDKPDVVRELIKLGAKPDLQEQYAGAATAASAAGMHPLDALQRREDEFCGGMNARASFLGWTALHYAALADSHDAAAALLDAGADPAARDHAGRRPLHYARDASPTRDLIQRAVRRREEEAAAAAAEERRRFPLERRLKQYIVGQRAAIHTVAAAVRRKENGWADDDHPLVFLFLGSSGIGKTELAKQLARYMHKDDPAAFIRLDMSEYQEKHEVAKLIGAPPGYVGHEEGGQLTRALAQRADAVVLFDEVDKAHPDVLTVLLQLFDEGRLTDGKGKLIECKDAIFVMTSNLAADEIAQYGLQLRREADALAATRRGKREGEEEEQEPTLEVSRHFKDNVVRPILKRQFRRDEFLGRINEIVYFLPFSRAELLALVQAELQRWAARARQRHGMLLLWEDDVLGALADGYEVHYGARSIQHEVERRVVSQLALAAERGLVARGCTLRLRAAPAGLALALRPAGAADFQPLHLPPA
Summary
Description
May function as a regulatory ATPase and be related to secretion/protein trafficking process.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the ClpA/ClpB family.
Keywords
Acetylation
ANK repeat
ATP-binding
Complete proteome
Direct protein sequencing
Hydrolase
Mitochondrion
Nucleotide-binding
Reference proteome
Repeat
Transit peptide
Alternative splicing
Cataract
Disease mutation
Epilepsy
Polymorphism
Feature
chain Caseinolytic peptidase B protein homolog
splice variant In isoform 5.
sequence variant In MEGCANN; dbSNP:rs200032855.
splice variant In isoform 5.
sequence variant In MEGCANN; dbSNP:rs200032855.
Uniprot
A0A194RF89
A0A3S2LAG8
A0A212EVL5
A0A2W1BVE1
A0A2H1WSK3
A0A194QCB5
+ More
D2A353 A0A0C9R1I9 E0W1P9 A0A1B6LFU2 A0A195F3L5 A0A2J7QRE5 A0A151X627 A0A151IHI6 A0A1W4WIW1 A0A1S3D5Y5 A0A232EU77 K7IY87 A0A1B6LS81 F4WH80 A0A452GHC6 A0A158NPE1 W4ZB95 B1H3I9 A0A1B6E1C0 A0A1W4WJ40 T1JLA4 Q3UGS9 E9ISR8 A0A195E5W1 A0A2C9JNP2 A0A087T6T7 A0A250YJT5 A0A154P6V3 A0A1Y1KPN5 A0A287DFJ1 E2ADV2 Q6IRH7 A0A340XS92 Q3U3U6 T1EDJ8 Q3URM1 Q60649 Q8CCN6 A0A2A3ETP3 A0A0P6J2T6 Q4R700 G3RM06 A0A0N0BG09 A0A2Y9F232 A0A2K5XK51 A0A0D9QVV8 Q3TXD4 Q5E9N5 A0A2K5X5G5 G2HEG2 F7A045 A0A2K6C1B4 A0A1S3A0Q8 A0A452SE80 A0A384DFC2 A0A2Y9DVC4 A0A2Y9I8X3 K7DLH6 W5P5H5 A0A452GA32 A0A2K6R666 A0A3Q0EFJ5 A0A2K6K9X5 U6CS37 A0A3Q7WLG7 A0A3Q7TM27 G3ICU8 A0A340XVA3 A0A1U7UIF0 A0A067R3L4 H0V6H2 Q9H078-2 A0A2Y9L846 A0A2I2ULQ1 G3U3C0 F6UA75 Q5RFS9 U3JUR8 F6XXW3 A0A1U7QVN4 A0A1S3HV43 A0A2U3VBC4 A0A2J8V1V7 A0A2K6GKN5 K9J2C5 A0A2K6UC39 F7G947 A0A3Q7QW97 A0A3Q7QWB0 A0A2Y9MWA4 W5MLW1 A0A3Q7S8C4 F7H9B9 A0A1S3HTQ1
D2A353 A0A0C9R1I9 E0W1P9 A0A1B6LFU2 A0A195F3L5 A0A2J7QRE5 A0A151X627 A0A151IHI6 A0A1W4WIW1 A0A1S3D5Y5 A0A232EU77 K7IY87 A0A1B6LS81 F4WH80 A0A452GHC6 A0A158NPE1 W4ZB95 B1H3I9 A0A1B6E1C0 A0A1W4WJ40 T1JLA4 Q3UGS9 E9ISR8 A0A195E5W1 A0A2C9JNP2 A0A087T6T7 A0A250YJT5 A0A154P6V3 A0A1Y1KPN5 A0A287DFJ1 E2ADV2 Q6IRH7 A0A340XS92 Q3U3U6 T1EDJ8 Q3URM1 Q60649 Q8CCN6 A0A2A3ETP3 A0A0P6J2T6 Q4R700 G3RM06 A0A0N0BG09 A0A2Y9F232 A0A2K5XK51 A0A0D9QVV8 Q3TXD4 Q5E9N5 A0A2K5X5G5 G2HEG2 F7A045 A0A2K6C1B4 A0A1S3A0Q8 A0A452SE80 A0A384DFC2 A0A2Y9DVC4 A0A2Y9I8X3 K7DLH6 W5P5H5 A0A452GA32 A0A2K6R666 A0A3Q0EFJ5 A0A2K6K9X5 U6CS37 A0A3Q7WLG7 A0A3Q7TM27 G3ICU8 A0A340XVA3 A0A1U7UIF0 A0A067R3L4 H0V6H2 Q9H078-2 A0A2Y9L846 A0A2I2ULQ1 G3U3C0 F6UA75 Q5RFS9 U3JUR8 F6XXW3 A0A1U7QVN4 A0A1S3HV43 A0A2U3VBC4 A0A2J8V1V7 A0A2K6GKN5 K9J2C5 A0A2K6UC39 F7G947 A0A3Q7QW97 A0A3Q7QWB0 A0A2Y9MWA4 W5MLW1 A0A3Q7S8C4 F7H9B9 A0A1S3HTQ1
EC Number
3.6.1.3
Pubmed
26354079
22118469
28756777
18362917
19820115
20566863
+ More
28648823 20075255 21719571 28562605 21347285 10349636 11042159 11076861 11217851 12466851 16141073 21282665 15562597 28087693 28004739 20798317 15489334 15057822 15632090 23254933 12040188 21183079 7835694 19468303 15944441 22398555 16305752 21484476 17431167 24813606 20809919 25362486 21804562 24845553 21993624 11230166 14702039 16554811 19608861 21269460 24275569 25944712 25597510 25597511 17975172 19892987 16341006 17495919 25243066
28648823 20075255 21719571 28562605 21347285 10349636 11042159 11076861 11217851 12466851 16141073 21282665 15562597 28087693 28004739 20798317 15489334 15057822 15632090 23254933 12040188 21183079 7835694 19468303 15944441 22398555 16305752 21484476 17431167 24813606 20809919 25362486 21804562 24845553 21993624 11230166 14702039 16554811 19608861 21269460 24275569 25944712 25597510 25597511 17975172 19892987 16341006 17495919 25243066
EMBL
KQ460297
KPJ16252.1
RSAL01000324
RVE42537.1
AGBW02012175
OWR45523.1
+ More
KZ149891 PZC79052.1 ODYU01010747 SOQ56050.1 KQ459439 KPJ01076.1 KQ971338 EFA01960.1 GBYB01001825 JAG71592.1 DS235873 EEB19631.1 GEBQ01017425 JAT22552.1 KQ981836 KYN35048.1 NEVH01011898 PNF31159.1 KQ982482 KYQ55855.1 KQ977643 KYN00863.1 NNAY01002164 OXU21902.1 GEBQ01013470 JAT26507.1 GL888153 EGI66446.1 ADTU01022402 AAGJ04144578 BC161411 AAI61411.1 GEDC01005575 JAS31723.1 JH431987 AK147772 BAE28128.1 GL765434 EFZ16386.1 KQ979608 KYN20272.1 KK113700 KFM60826.1 GFFW01000992 JAV43796.1 KQ434827 KZC07553.1 GEZM01081464 JAV61625.1 AGTP01028809 AGTP01028810 GL438820 EFN68464.1 AABR07004904 AABR07071951 AC122631 BC070917 CH473956 AAH70917.1 EDM18274.1 AK154577 BAE32689.1 AMQM01007880 KB097680 ESN91999.1 AK138593 AK141383 CH466531 BAE23708.1 BAE24667.1 EDL16519.1 U09874 BC048175 AC127586 AC129609 AC159005 AK032421 BAC27861.1 KZ288189 PBC34652.1 GEBF01007007 JAN96625.1 AB169030 BAE01124.1 CABD030080096 CABD030080097 CABD030080098 CABD030080099 CABD030080100 CABD030080101 CABD030080102 CABD030080103 CABD030080104 CABD030080105 KQ435794 KOX73872.1 AQIB01134177 AQIB01134178 AK159314 BAE34982.1 BT020885 BC103469 AQIA01021325 AQIA01021326 AQIA01021327 AQIA01021328 AQIA01021329 AK305126 BAK62120.1 JSUE03012881 GABE01008702 NBAG03000097 JAA36037.1 PNI89519.1 AMGL01032362 AMGL01032363 LWLT01000015 HAAF01001308 CCP73134.1 JH001963 EGW14086.1 KK852727 KDR17615.1 AAKN02051221 AAKN02051222 AAKN02051223 AAKN02051224 AL136909 AL834484 AK023214 AK302006 AK302069 AK302158 AP000593 AP002892 AP003785 BC006404 AANG04002890 CR857073 CAH89378.1 AGTO01011075 AAEX03012823 AAEX03012824 AAEX03012825 NDHI03003437 PNJ51507.1 GABZ01004826 JAA48699.1 AHAT01003026 AHAT01003027 GAMT01000493 GAMS01009293 GAMR01009336 GAMQ01000541 GAMP01001587 JAB11368.1 JAB13843.1 JAB24596.1 JAB41310.1 JAB51168.1
KZ149891 PZC79052.1 ODYU01010747 SOQ56050.1 KQ459439 KPJ01076.1 KQ971338 EFA01960.1 GBYB01001825 JAG71592.1 DS235873 EEB19631.1 GEBQ01017425 JAT22552.1 KQ981836 KYN35048.1 NEVH01011898 PNF31159.1 KQ982482 KYQ55855.1 KQ977643 KYN00863.1 NNAY01002164 OXU21902.1 GEBQ01013470 JAT26507.1 GL888153 EGI66446.1 ADTU01022402 AAGJ04144578 BC161411 AAI61411.1 GEDC01005575 JAS31723.1 JH431987 AK147772 BAE28128.1 GL765434 EFZ16386.1 KQ979608 KYN20272.1 KK113700 KFM60826.1 GFFW01000992 JAV43796.1 KQ434827 KZC07553.1 GEZM01081464 JAV61625.1 AGTP01028809 AGTP01028810 GL438820 EFN68464.1 AABR07004904 AABR07071951 AC122631 BC070917 CH473956 AAH70917.1 EDM18274.1 AK154577 BAE32689.1 AMQM01007880 KB097680 ESN91999.1 AK138593 AK141383 CH466531 BAE23708.1 BAE24667.1 EDL16519.1 U09874 BC048175 AC127586 AC129609 AC159005 AK032421 BAC27861.1 KZ288189 PBC34652.1 GEBF01007007 JAN96625.1 AB169030 BAE01124.1 CABD030080096 CABD030080097 CABD030080098 CABD030080099 CABD030080100 CABD030080101 CABD030080102 CABD030080103 CABD030080104 CABD030080105 KQ435794 KOX73872.1 AQIB01134177 AQIB01134178 AK159314 BAE34982.1 BT020885 BC103469 AQIA01021325 AQIA01021326 AQIA01021327 AQIA01021328 AQIA01021329 AK305126 BAK62120.1 JSUE03012881 GABE01008702 NBAG03000097 JAA36037.1 PNI89519.1 AMGL01032362 AMGL01032363 LWLT01000015 HAAF01001308 CCP73134.1 JH001963 EGW14086.1 KK852727 KDR17615.1 AAKN02051221 AAKN02051222 AAKN02051223 AAKN02051224 AL136909 AL834484 AK023214 AK302006 AK302069 AK302158 AP000593 AP002892 AP003785 BC006404 AANG04002890 CR857073 CAH89378.1 AGTO01011075 AAEX03012823 AAEX03012824 AAEX03012825 NDHI03003437 PNJ51507.1 GABZ01004826 JAA48699.1 AHAT01003026 AHAT01003027 GAMT01000493 GAMS01009293 GAMR01009336 GAMQ01000541 GAMP01001587 JAB11368.1 JAB13843.1 JAB24596.1 JAB41310.1 JAB51168.1
Proteomes
UP000053240
UP000283053
UP000007151
UP000053268
UP000007266
UP000009046
+ More
UP000078541 UP000235965 UP000075809 UP000078542 UP000192223 UP000079169 UP000215335 UP000002358 UP000007755 UP000291020 UP000005205 UP000007110 UP000078492 UP000076420 UP000054359 UP000076502 UP000005215 UP000000311 UP000002494 UP000265300 UP000015101 UP000000589 UP000242457 UP000001519 UP000053105 UP000248484 UP000233140 UP000029965 UP000009136 UP000233100 UP000006718 UP000233120 UP000079721 UP000291022 UP000261680 UP000291021 UP000248480 UP000248481 UP000002356 UP000291000 UP000233200 UP000189704 UP000233180 UP000286642 UP000286640 UP000001075 UP000027135 UP000005447 UP000005640 UP000248482 UP000011712 UP000007646 UP000002281 UP000016665 UP000002254 UP000189706 UP000085678 UP000245340 UP000233160 UP000233220 UP000002280 UP000286641 UP000248483 UP000018468 UP000008225
UP000078541 UP000235965 UP000075809 UP000078542 UP000192223 UP000079169 UP000215335 UP000002358 UP000007755 UP000291020 UP000005205 UP000007110 UP000078492 UP000076420 UP000054359 UP000076502 UP000005215 UP000000311 UP000002494 UP000265300 UP000015101 UP000000589 UP000242457 UP000001519 UP000053105 UP000248484 UP000233140 UP000029965 UP000009136 UP000233100 UP000006718 UP000233120 UP000079721 UP000291022 UP000261680 UP000291021 UP000248480 UP000248481 UP000002356 UP000291000 UP000233200 UP000189704 UP000233180 UP000286642 UP000286640 UP000001075 UP000027135 UP000005447 UP000005640 UP000248482 UP000011712 UP000007646 UP000002281 UP000016665 UP000002254 UP000189706 UP000085678 UP000245340 UP000233160 UP000233220 UP000002280 UP000286641 UP000248483 UP000018468 UP000008225
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194RF89
A0A3S2LAG8
A0A212EVL5
A0A2W1BVE1
A0A2H1WSK3
A0A194QCB5
+ More
D2A353 A0A0C9R1I9 E0W1P9 A0A1B6LFU2 A0A195F3L5 A0A2J7QRE5 A0A151X627 A0A151IHI6 A0A1W4WIW1 A0A1S3D5Y5 A0A232EU77 K7IY87 A0A1B6LS81 F4WH80 A0A452GHC6 A0A158NPE1 W4ZB95 B1H3I9 A0A1B6E1C0 A0A1W4WJ40 T1JLA4 Q3UGS9 E9ISR8 A0A195E5W1 A0A2C9JNP2 A0A087T6T7 A0A250YJT5 A0A154P6V3 A0A1Y1KPN5 A0A287DFJ1 E2ADV2 Q6IRH7 A0A340XS92 Q3U3U6 T1EDJ8 Q3URM1 Q60649 Q8CCN6 A0A2A3ETP3 A0A0P6J2T6 Q4R700 G3RM06 A0A0N0BG09 A0A2Y9F232 A0A2K5XK51 A0A0D9QVV8 Q3TXD4 Q5E9N5 A0A2K5X5G5 G2HEG2 F7A045 A0A2K6C1B4 A0A1S3A0Q8 A0A452SE80 A0A384DFC2 A0A2Y9DVC4 A0A2Y9I8X3 K7DLH6 W5P5H5 A0A452GA32 A0A2K6R666 A0A3Q0EFJ5 A0A2K6K9X5 U6CS37 A0A3Q7WLG7 A0A3Q7TM27 G3ICU8 A0A340XVA3 A0A1U7UIF0 A0A067R3L4 H0V6H2 Q9H078-2 A0A2Y9L846 A0A2I2ULQ1 G3U3C0 F6UA75 Q5RFS9 U3JUR8 F6XXW3 A0A1U7QVN4 A0A1S3HV43 A0A2U3VBC4 A0A2J8V1V7 A0A2K6GKN5 K9J2C5 A0A2K6UC39 F7G947 A0A3Q7QW97 A0A3Q7QWB0 A0A2Y9MWA4 W5MLW1 A0A3Q7S8C4 F7H9B9 A0A1S3HTQ1
D2A353 A0A0C9R1I9 E0W1P9 A0A1B6LFU2 A0A195F3L5 A0A2J7QRE5 A0A151X627 A0A151IHI6 A0A1W4WIW1 A0A1S3D5Y5 A0A232EU77 K7IY87 A0A1B6LS81 F4WH80 A0A452GHC6 A0A158NPE1 W4ZB95 B1H3I9 A0A1B6E1C0 A0A1W4WJ40 T1JLA4 Q3UGS9 E9ISR8 A0A195E5W1 A0A2C9JNP2 A0A087T6T7 A0A250YJT5 A0A154P6V3 A0A1Y1KPN5 A0A287DFJ1 E2ADV2 Q6IRH7 A0A340XS92 Q3U3U6 T1EDJ8 Q3URM1 Q60649 Q8CCN6 A0A2A3ETP3 A0A0P6J2T6 Q4R700 G3RM06 A0A0N0BG09 A0A2Y9F232 A0A2K5XK51 A0A0D9QVV8 Q3TXD4 Q5E9N5 A0A2K5X5G5 G2HEG2 F7A045 A0A2K6C1B4 A0A1S3A0Q8 A0A452SE80 A0A384DFC2 A0A2Y9DVC4 A0A2Y9I8X3 K7DLH6 W5P5H5 A0A452GA32 A0A2K6R666 A0A3Q0EFJ5 A0A2K6K9X5 U6CS37 A0A3Q7WLG7 A0A3Q7TM27 G3ICU8 A0A340XVA3 A0A1U7UIF0 A0A067R3L4 H0V6H2 Q9H078-2 A0A2Y9L846 A0A2I2ULQ1 G3U3C0 F6UA75 Q5RFS9 U3JUR8 F6XXW3 A0A1U7QVN4 A0A1S3HV43 A0A2U3VBC4 A0A2J8V1V7 A0A2K6GKN5 K9J2C5 A0A2K6UC39 F7G947 A0A3Q7QW97 A0A3Q7QWB0 A0A2Y9MWA4 W5MLW1 A0A3Q7S8C4 F7H9B9 A0A1S3HTQ1
PDB
4D2X
E-value=4.50233e-48,
Score=484
Ontologies
GO
Topology
Subcellular location
Mitochondrion
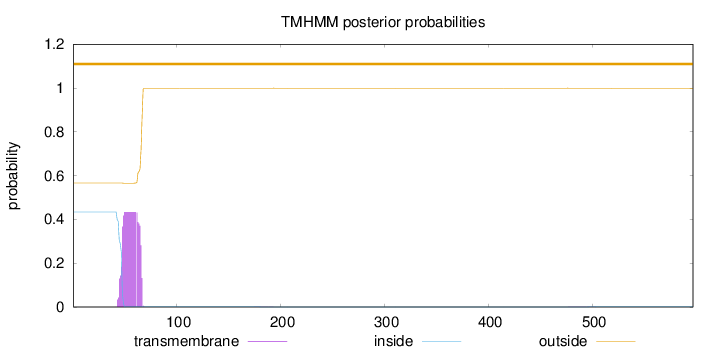
Length:
597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.52441999999998
Exp number, first 60 AAs:
6.08968
Total prob of N-in:
0.43411
outside
1 - 597
Population Genetic Test Statistics
Pi
3.719428
Theta
7.3751
Tajima's D
-1.648653
CLR
38.785824
CSRT
0.0380980950952452
Interpretation
Uncertain
