Gene
KWMTBOMO08138
Pre Gene Modal
BGIBMGA001075
Annotation
PREDICTED:_phosphatidylcholine:ceramide_cholinephosphotransferase_2-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.677
Sequence
CDS
ATGACGATCGCAGCACGCTCCCCAGATAAGGAGCCGTCGCCACCAAAACAAATTAACTTCGCCGAGCCGCTAGTGGAGTCTGTGCGTGTGTATCTCGCAAATGAAGTGCCTTCCAAGCAGCTAGCCCTCGTGCCCTGCGTGCCCGCGTGCCGGCCCTCGCCCCCGCCGACTCCTCCCGTCTGTGATGTCGACATAGCCTCTCAGACCGCTCGCGAAGAGATCAAGGGAATCAACTTCGAGGCGCGCTTCAAACGGAATGACCTAAACAACAATCCTCCTCGCCAACAAAACGACCAACCACAAAACGGCACCATGCCCAGCCAAGCGGAACTCATGCAGAGGCAACCGCTCTTGGCCCGCACCACCAAAACAGACGCACCGCAGCCAGCCACGGACGAGTCCGACCCTGACCAGGACCACTCTCCGCAGAGAACGCACACCGGCGACTTCATCATAGAGTTACCTCCGGGCACCGTCAGAGAAGAAAGATACCCTAAAGAAATATGGAAAACGATTCTATCATTTTTCTTCCTATTCATGTGCGTTTGCATCAACATGATTTCACTATCGCTGGTGCACGAGCGACTGCCGGACCGGAACACGACGAAGCCGCTCAACGACATAGTGTTAGACAACGTGCCGGCGCACGACTGGGGCCTGGCGGTCTCCGAGTACCTGATCATGGTGTCAACTACGGTGGCGATGCTAGTGGTCGTGTTCCACAAGCACCGCTTCATCGTGGCGCGTCGGCTGTTCTTCATCATCGGCCTGCTGTACTTGTACCGCTCGATCACGATGTTCGTGACCGTGTTGCCGGTGTCCAGCATGACGTACACCTGTAGCCCAAAGAGCAACAGCACGGCCCCGCTGCTCATCATCAAGAGAATGTTCTACTTGATATCCGGCTTCGGGCTCTCGATAAACGGAAAACACACATACTGTGGAGACTACATATACTCGGGGCACACGATGGTGTTGGTGCTCAGCTATCTGATAGTCGCGGAATACTCACCCAAGAAGTTGTGGCCAATGCACTGGGCGATGGCGGGCTCCGCCGTGATGGGCGTCTCGTTCGTCCTGCTCGCCCACGGTCACTACACCGTGGACGTGGAAACGGATACAACCATTACATGA
Protein
MTIAARSPDKEPSPPKQINFAEPLVESVRVYLANEVPSKQLALVPCVPACRPSPPPTPPVCDVDIASQTAREEIKGINFEARFKRNDLNNNPPRQQNDQPQNGTMPSQAELMQRQPLLARTTKTDAPQPATDESDPDQDHSPQRTHTGDFIIELPPGTVREERYPKEIWKTILSFFFLFMCVCINMISLSLVHERLPDRNTTKPLNDIVLDNVPAHDWGLAVSEYLIMVSTTVAMLVVVFHKHRFIVARRLFFIIGLLYLYRSITMFVTVLPVSSMTYTCSPKSNSTAPLLIIKRMFYLISGFGLSINGKHTYCGDYIYSGHTMVLVLSYLIVAEYSPKKLWPMHWAMAGSAVMGVSFVLLAHGHYTVDVETDTTIT
Summary
Uniprot
H9IUZ5
A0A2H1WSV0
A0A2A4J8T3
A0A2W1C4I3
A0A194Q6B4
A0A194RJX4
+ More
A0A0M8ZQK7 V9I9T6 A0A2A3EDQ8 V9IB60 A0A0L7RGZ2 E2AM13 A0A139W971 A0A139W966 K7J605 A0A026W8W8 A0A154PCZ4 A0A088AN88 A0A1B6C4E3 A0A1B6C209 A0A1B6E5A4 A0A0C9RA80 A0A0C9QNL7 A0A1W4WWY5 V5GRY5 E0VSX9 A0A2J7PY50 A0A224XHQ6 A0A023F4W2 A0A067QXD3 A0A146LG08 A0A0A9Z724 A0A0A9W2M5 A0A0A9XSF9 T1HMV0 A0A0K8SX40 A0A0A9YCX7 A0A0K8SXG5
A0A0M8ZQK7 V9I9T6 A0A2A3EDQ8 V9IB60 A0A0L7RGZ2 E2AM13 A0A139W971 A0A139W966 K7J605 A0A026W8W8 A0A154PCZ4 A0A088AN88 A0A1B6C4E3 A0A1B6C209 A0A1B6E5A4 A0A0C9RA80 A0A0C9QNL7 A0A1W4WWY5 V5GRY5 E0VSX9 A0A2J7PY50 A0A224XHQ6 A0A023F4W2 A0A067QXD3 A0A146LG08 A0A0A9Z724 A0A0A9W2M5 A0A0A9XSF9 T1HMV0 A0A0K8SX40 A0A0A9YCX7 A0A0K8SXG5
Pubmed
EMBL
BABH01000043
ODYU01010747
SOQ56052.1
NWSH01002346
PCG68475.1
KZ149891
+ More
PZC79053.1 KQ459439 KPJ01073.1 KQ460297 KPJ16251.1 KQ436008 KOX67623.1 JR037636 AEY57883.1 KZ288292 PBC29151.1 JR037635 AEY57882.1 KQ414596 KOC69961.1 GL440703 EFN65533.1 KQ972625 KXZ75838.1 KXZ75837.1 KK107341 EZA52428.1 KQ434878 KZC09799.1 GEDC01028941 JAS08357.1 GEDC01029999 GEDC01001733 JAS07299.1 JAS35565.1 GEDC01004186 JAS33112.1 GBYB01005163 GBYB01005165 JAG74930.1 JAG74932.1 GBYB01005164 JAG74931.1 GALX01004069 JAB64397.1 DS235758 EEB16485.1 NEVH01020850 PNF21261.1 GFTR01005852 JAW10574.1 GBBI01002649 JAC16063.1 KK853245 KDR09403.1 GDHC01011561 JAQ07068.1 GBHO01041945 GBHO01041940 GBHO01041218 GBHO01041217 GBHO01041216 GBHO01041215 GBHO01041214 GBHO01041213 GBHO01023799 GBHO01003908 GDHC01015134 JAG01659.1 JAG01664.1 JAG02386.1 JAG02387.1 JAG02388.1 JAG02389.1 JAG02390.1 JAG02391.1 JAG19805.1 JAG39696.1 JAQ03495.1 GBHO01041943 GBHO01041220 GBHO01023801 GBHO01012669 GBHO01012666 GBRD01007953 GDHC01014711 GDHC01012960 JAG01661.1 JAG02384.1 JAG19803.1 JAG30935.1 JAG30938.1 JAG57868.1 JAQ03918.1 JAQ05669.1 GBHO01041212 GBHO01021018 GBHO01012671 GBHO01012670 GBHO01012668 GDHC01008049 JAG02392.1 JAG22586.1 JAG30933.1 JAG30934.1 JAG30936.1 JAQ10580.1 ACPB03008518 GBRD01007954 JAG57867.1 GBHO01041219 GBHO01041211 GBHO01023797 GBHO01012672 GBHO01012667 JAG02385.1 JAG02393.1 JAG19807.1 JAG30932.1 JAG30937.1 GBRD01007955 JAG57866.1
PZC79053.1 KQ459439 KPJ01073.1 KQ460297 KPJ16251.1 KQ436008 KOX67623.1 JR037636 AEY57883.1 KZ288292 PBC29151.1 JR037635 AEY57882.1 KQ414596 KOC69961.1 GL440703 EFN65533.1 KQ972625 KXZ75838.1 KXZ75837.1 KK107341 EZA52428.1 KQ434878 KZC09799.1 GEDC01028941 JAS08357.1 GEDC01029999 GEDC01001733 JAS07299.1 JAS35565.1 GEDC01004186 JAS33112.1 GBYB01005163 GBYB01005165 JAG74930.1 JAG74932.1 GBYB01005164 JAG74931.1 GALX01004069 JAB64397.1 DS235758 EEB16485.1 NEVH01020850 PNF21261.1 GFTR01005852 JAW10574.1 GBBI01002649 JAC16063.1 KK853245 KDR09403.1 GDHC01011561 JAQ07068.1 GBHO01041945 GBHO01041940 GBHO01041218 GBHO01041217 GBHO01041216 GBHO01041215 GBHO01041214 GBHO01041213 GBHO01023799 GBHO01003908 GDHC01015134 JAG01659.1 JAG01664.1 JAG02386.1 JAG02387.1 JAG02388.1 JAG02389.1 JAG02390.1 JAG02391.1 JAG19805.1 JAG39696.1 JAQ03495.1 GBHO01041943 GBHO01041220 GBHO01023801 GBHO01012669 GBHO01012666 GBRD01007953 GDHC01014711 GDHC01012960 JAG01661.1 JAG02384.1 JAG19803.1 JAG30935.1 JAG30938.1 JAG57868.1 JAQ03918.1 JAQ05669.1 GBHO01041212 GBHO01021018 GBHO01012671 GBHO01012670 GBHO01012668 GDHC01008049 JAG02392.1 JAG22586.1 JAG30933.1 JAG30934.1 JAG30936.1 JAQ10580.1 ACPB03008518 GBRD01007954 JAG57867.1 GBHO01041219 GBHO01041211 GBHO01023797 GBHO01012672 GBHO01012667 JAG02385.1 JAG02393.1 JAG19807.1 JAG30932.1 JAG30937.1 GBRD01007955 JAG57866.1
Proteomes
PRIDE
Pfam
PF14360 PAP2_C
Interpro
IPR025749
Sphingomyelin_synth-like_dom
ProteinModelPortal
H9IUZ5
A0A2H1WSV0
A0A2A4J8T3
A0A2W1C4I3
A0A194Q6B4
A0A194RJX4
+ More
A0A0M8ZQK7 V9I9T6 A0A2A3EDQ8 V9IB60 A0A0L7RGZ2 E2AM13 A0A139W971 A0A139W966 K7J605 A0A026W8W8 A0A154PCZ4 A0A088AN88 A0A1B6C4E3 A0A1B6C209 A0A1B6E5A4 A0A0C9RA80 A0A0C9QNL7 A0A1W4WWY5 V5GRY5 E0VSX9 A0A2J7PY50 A0A224XHQ6 A0A023F4W2 A0A067QXD3 A0A146LG08 A0A0A9Z724 A0A0A9W2M5 A0A0A9XSF9 T1HMV0 A0A0K8SX40 A0A0A9YCX7 A0A0K8SXG5
A0A0M8ZQK7 V9I9T6 A0A2A3EDQ8 V9IB60 A0A0L7RGZ2 E2AM13 A0A139W971 A0A139W966 K7J605 A0A026W8W8 A0A154PCZ4 A0A088AN88 A0A1B6C4E3 A0A1B6C209 A0A1B6E5A4 A0A0C9RA80 A0A0C9QNL7 A0A1W4WWY5 V5GRY5 E0VSX9 A0A2J7PY50 A0A224XHQ6 A0A023F4W2 A0A067QXD3 A0A146LG08 A0A0A9Z724 A0A0A9W2M5 A0A0A9XSF9 T1HMV0 A0A0K8SX40 A0A0A9YCX7 A0A0K8SXG5
Ontologies
PATHWAY
GO
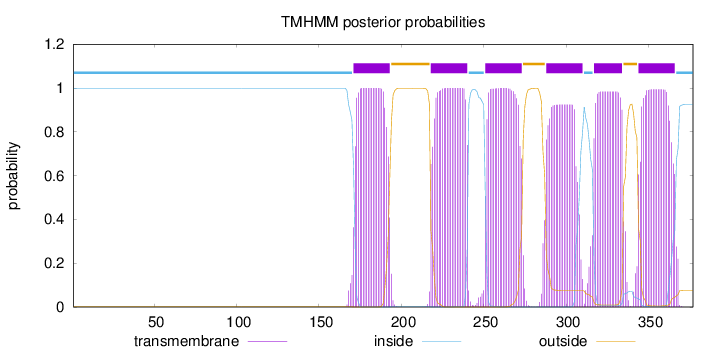
Topology
Length:
377
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
125.48639
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99928
inside
1 - 170
TMhelix
171 - 193
outside
194 - 217
TMhelix
218 - 240
inside
241 - 250
TMhelix
251 - 273
outside
274 - 287
TMhelix
288 - 310
inside
311 - 316
TMhelix
317 - 334
outside
335 - 343
TMhelix
344 - 366
inside
367 - 377
Population Genetic Test Statistics
Pi
21.187016
Theta
25.364846
Tajima's D
-0.494279
CLR
5.871875
CSRT
0.242937853107345
Interpretation
Uncertain
