Gene
KWMTBOMO08135 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001091
Annotation
PREDICTED:_spondin-1-like_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 2.688
Sequence
CDS
ATGGGCAATCTGAGAATTTTAGTGTGGCTGGGCCTAGTGTCCGCGGCACTGGCCTGCGAGCTAAATCCAGGTCCTGGAGTAGGTTCAAAGTCACCTGGAGACAATCACTACAGGCTGATTGTTAACGGAGAAGTTGAGCGATATGCGCCGGATCAGAGATATGTCGTGACTCTGGTCGGTTCCCGCACTCATGACGTAGTACAACAGTTTGCGGGCTTCAAAATAATCCTTGATCCTTTGAACCCGGACACAAGGAGGGCGCCCAGCAAACAGGGACAGTTCCAGCTCTTCGCCGATACTTTAACTAAGTTCGACGAAGAATGTACGAACTCCGTTGTCGAGGCTGACGATCTGCCCAAAACTGAAGTCCAGGTCATGTGGAAAGCGCCACCGGCTGGTTCAGGATGCGTGCTTTTGAAGGCGATGGTGTATGAAAATGCGAGTCGTTGGTTCGCTGAAGACGGTCAGTTGACCAAACGAATCTGCGAGGACACGTCTCTGTCCATCCCCGACTGCTGCGCCTGCGACGACGCTAAATATAGGATGGTGTTCGAGGGGCTCTGGTCTCCGCAGACACATCCAAAGAACTTCCCGACCCAAGCTCTGTGGCTAACTCACTTCTCTGACGTCATCGGAGCCACTCATCCTAAGAATTTCACTTTCTGGGGCGAAGGACAGCTCGCTTCAGACGGATTTAGATCTCTGGCGGAATGGGGTTCAGTAGGTCTGATGGAGCGTGAACTTCGTCAACACGGAGGTCTGCTCCGGTCCATCGTCAAGGCCCAGGGTCTGTGGCATCCGAGGGTCAATTCCAATACTTCTGCTGCCTTCACTGTTGACAAAAAGAGGCATTACCTCTCTCTTGCTTCTATGTTTGGTCCATCTCCTGACTGGGTGGTGGGAGTGAGTGGACTGGACCTCTGCCAGAAAGATTGCTCCTGGGTTGAATCTAAGATTATCGACTTGTATCCTTACGACGCTGGCACAGACAATGGCGTCTCTTACATGTCGCCCAATTCTGAGACCGTACCCCGGGAAAGGATGTACCGCATCACCCCCATGTTCCCCGAGGACCCTCGTGCTCCGTTCTACGACCCCGACTCCAAGACGATGGCTCCGATGGCCAGACTCTACCTCACCAGAGAAAAATTAATTTCCAAATCTTGTGACGAGGAAACTCTTTTAGCTCTCGTCGCTGAAGAGGAAGAGAACACGCAGACGGTCGATAAACCGCAATGTGCGGTGACGGAGTGGAGCGCGTGGTCGGAGTGCTCCGTGAGCTGCGGCAAGGGGCTCCGCATGAGGACGCGCCAGTACCGCCTGCCCGACAAGGCGCGCATGTTCTCCTGCGACCGCCAGCTCGTCTCCAAGGAGATGTGCGTCGCGCCCGTCGCCGAGTGCGACGGTGACGGCGAAGGTGACTCGGAGGCCGACAGCGTGGGCACCCCGGTGGAGGACCTGGACGGAGTGTGCAAGACCTACGACTGGGGCTCGTGGAGCGAGTGCTCCGTCACGTGCGGCGTCGGCATCAGCACCAGGAGACGACAGTTCGTCAACCACATGGGACTGAAGAAATGTCCGCTTGTGCAGATCGAAGAGAACCGCAAGTGCATGGAGCCGGCGTGCGCGGAGGAGGAGGCGGCGGCGGCGGCGGACCCGCGCTGCCCCACGTCGGCGTGGTCGGGCTGGTCGCCGTGCTCGGCGTCGTGCGGGCGCGGGGTGCGCTTCCGCACGCGGCTGCTGCTGGTGCCGGCCGACCAGCAGGCCGCGTGCGCCGCCGCCGTGGAGCTGCTGCAGCAGCGGCCCTGCTCCGACCGGGACGACTGCACCATCGACATGCTCACCGCCAAACGCATATGTATGGAGGAACCCACTCAAGGGCCGTGCAGAGGACTGTACCAGCGCTGGGCCTTCGTGGCCATGAAGGGGATGTGCATTCCCTTCAACTACGGCGGATGTCGCGGCAGCCAGAACAACTTCCTCACGCAAGAAGACTGTATGAACACTTGCAAGATTATGCTCGGTGGAGTAGTGCCGGGGGGATCAGTGCCTTCGCCGGCACTTCTACCGTCGCCCGTCGTCAGTTCCAACTATCCAGGACTCAGTATCAGCTCCATTGTGCCTCTTTCTAACGGTGTTGGTGGTCCTACCCTGACGGGAAGTCCAGGAGACTGTCAGGTGAGCCAGTGGAGCGACTGGAGCAAGTGCAGCCAGCCCTGCGGAGTCGGCTACCAGGAGCGCGTCAGGACTATTCTTGCACAGCCCGGCCCGGGAGGAGTGCCGTGTCCGAAGCTAATGTCGCGGCGCCGACGGTGCTTCAGGCGGTGTTAA
Protein
MGNLRILVWLGLVSAALACELNPGPGVGSKSPGDNHYRLIVNGEVERYAPDQRYVVTLVGSRTHDVVQQFAGFKIILDPLNPDTRRAPSKQGQFQLFADTLTKFDEECTNSVVEADDLPKTEVQVMWKAPPAGSGCVLLKAMVYENASRWFAEDGQLTKRICEDTSLSIPDCCACDDAKYRMVFEGLWSPQTHPKNFPTQALWLTHFSDVIGATHPKNFTFWGEGQLASDGFRSLAEWGSVGLMERELRQHGGLLRSIVKAQGLWHPRVNSNTSAAFTVDKKRHYLSLASMFGPSPDWVVGVSGLDLCQKDCSWVESKIIDLYPYDAGTDNGVSYMSPNSETVPRERMYRITPMFPEDPRAPFYDPDSKTMAPMARLYLTREKLISKSCDEETLLALVAEEEENTQTVDKPQCAVTEWSAWSECSVSCGKGLRMRTRQYRLPDKARMFSCDRQLVSKEMCVAPVAECDGDGEGDSEADSVGTPVEDLDGVCKTYDWGSWSECSVTCGVGISTRRRQFVNHMGLKKCPLVQIEENRKCMEPACAEEEAAAAADPRCPTSAWSGWSPCSASCGRGVRFRTRLLLVPADQQAACAAAVELLQQRPCSDRDDCTIDMLTAKRICMEEPTQGPCRGLYQRWAFVAMKGMCIPFNYGGCRGSQNNFLTQEDCMNTCKIMLGGVVPGGSVPSPALLPSPVVSSNYPGLSISSIVPLSNGVGGPTLTGSPGDCQVSQWSDWSKCSQPCGVGYQERVRTILAQPGPGGVPCPKLMSRRRRCFRRC
Summary
Uniprot
H9IV11
A0A2A4JL45
A0A2H1WUG2
A0A212EVP3
A0A194REI6
A0A2W1C015
+ More
U5ETX1 A0A182X6R7 D2A031 A0A182HH71 A0A182US71 A0A067QSB9 A0A182W1R7 A0A084WJ48 A0A182IWE1 Q7PZ75 A0A182FW84 A0A182H1I6 A0A2J7PZ95 A0A182L4P3 A0A182N3K1 A0A1Y1K2N4 A0A182RVE0 A0A182MDJ6 B0WRZ7 A0A182PSA0 A0A2A3ES07 A0A154NXF1 A0A1Q3FLK1 W5JTW2 A0A088AUJ4 A0A182KA48 Q17FH5 A0A182G628 V5H0T5 A0A2M4A123 A0A1S4F4R7 E2AZK3 A0A2M3YYU8 A0A2M4A1L1 A0A2M4A1B5 A0A2M4BEF7 A0A2M3YZ25 A0A2M4A1T8 A0A1W4XFH7 A0A182QMH4 A0A182TJA2 J3JTM0 T1EA54 E2BGT3 T1PNB9 A0A1I8MX43 A0A1A9UVC8 A0A026W2S5 A0A1B0ACU2 A0A1I8PHY5 A0A1A9Y5X7 A0A195F377 A0A1B0B2H8 A0A0L0CJP3 A0A2M4BFK2 A0A195BSB4 A0A158P1Q3 Q290S6 B4GBA8 A0A1A9WIB2 A0A1B0FAY9 W8BN93 B4JVZ6 A0A336KJF1 A0A0L7RKC0 A0A1J1HR27 A0A1W4VBY5 B4LJC8 A0A3B0JJI0 A0A1B6CP24 A0A0K8U3T0 B3MD67 B4N5T5 Q7KN04 A0A0A1WWX3 B4QIJ8 B4P8H9 A0A195D1Y4 A0A411G7Z9 A0A151WRA0 B4HMH1 B4KTS4 A0A232ELX5 E0W462 K7IZ87 A0A0M4ESX8 B3NP25 A0A034V5Z7 A0A182YL14 A0A0K8V430 A0A0A1XE95 F4WMF1 T1PLM0 N6TGR9
U5ETX1 A0A182X6R7 D2A031 A0A182HH71 A0A182US71 A0A067QSB9 A0A182W1R7 A0A084WJ48 A0A182IWE1 Q7PZ75 A0A182FW84 A0A182H1I6 A0A2J7PZ95 A0A182L4P3 A0A182N3K1 A0A1Y1K2N4 A0A182RVE0 A0A182MDJ6 B0WRZ7 A0A182PSA0 A0A2A3ES07 A0A154NXF1 A0A1Q3FLK1 W5JTW2 A0A088AUJ4 A0A182KA48 Q17FH5 A0A182G628 V5H0T5 A0A2M4A123 A0A1S4F4R7 E2AZK3 A0A2M3YYU8 A0A2M4A1L1 A0A2M4A1B5 A0A2M4BEF7 A0A2M3YZ25 A0A2M4A1T8 A0A1W4XFH7 A0A182QMH4 A0A182TJA2 J3JTM0 T1EA54 E2BGT3 T1PNB9 A0A1I8MX43 A0A1A9UVC8 A0A026W2S5 A0A1B0ACU2 A0A1I8PHY5 A0A1A9Y5X7 A0A195F377 A0A1B0B2H8 A0A0L0CJP3 A0A2M4BFK2 A0A195BSB4 A0A158P1Q3 Q290S6 B4GBA8 A0A1A9WIB2 A0A1B0FAY9 W8BN93 B4JVZ6 A0A336KJF1 A0A0L7RKC0 A0A1J1HR27 A0A1W4VBY5 B4LJC8 A0A3B0JJI0 A0A1B6CP24 A0A0K8U3T0 B3MD67 B4N5T5 Q7KN04 A0A0A1WWX3 B4QIJ8 B4P8H9 A0A195D1Y4 A0A411G7Z9 A0A151WRA0 B4HMH1 B4KTS4 A0A232ELX5 E0W462 K7IZ87 A0A0M4ESX8 B3NP25 A0A034V5Z7 A0A182YL14 A0A0K8V430 A0A0A1XE95 F4WMF1 T1PLM0 N6TGR9
Pubmed
19121390
22118469
26354079
28756777
18362917
19820115
+ More
24845553 24438588 12364791 14747013 17210077 26483478 20966253 28004739 20920257 23761445 17510324 20798317 22516182 25315136 24508170 30249741 26108605 21347285 15632085 17994087 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22936249 17550304 28648823 20566863 20075255 25348373 25244985 21719571 23537049
24845553 24438588 12364791 14747013 17210077 26483478 20966253 28004739 20920257 23761445 17510324 20798317 22516182 25315136 24508170 30249741 26108605 21347285 15632085 17994087 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22936249 17550304 28648823 20566863 20075255 25348373 25244985 21719571 23537049
EMBL
BABH01000051
NWSH01001205
PCG72130.1
ODYU01010747
SOQ56054.1
AGBW02012175
+ More
OWR45527.1 KQ460297 KPJ16248.1 KZ149891 PZC79055.1 GANO01002591 JAB57280.1 KQ971338 EFA01753.1 APCN01002391 KK853014 KDR12501.1 ATLV01023973 KE525347 KFB50242.1 AAAB01008986 EAA00407.4 JXUM01023161 JXUM01023162 JXUM01023163 KQ560653 KXJ81428.1 NEVH01020337 PNF21664.1 GEZM01098931 JAV53636.1 AXCM01006417 DS232063 EDS33615.1 KZ288192 PBC34294.1 KQ434778 KZC04355.1 GFDL01006630 JAV28415.1 ADMH02000153 ETN67571.1 CH477271 EAT45270.1 JXUM01044332 JXUM01044333 KQ561396 KXJ78714.1 GALX01002028 JAB66438.1 GGFK01001193 MBW34514.1 GL444248 EFN61136.1 GGFM01000676 MBW21427.1 GGFK01001207 MBW34528.1 GGFK01001214 MBW34535.1 GGFJ01002281 MBW51422.1 GGFM01000740 MBW21491.1 GGFK01001455 MBW34776.1 AXCN02001106 BT126577 AEE61541.1 GAMD01001308 JAB00283.1 GL448228 EFN85065.1 KA649560 AFP64189.1 KK107488 QOIP01000012 EZA49891.1 RLU16301.1 KQ981855 KYN34836.1 JXJN01007649 JRES01000299 KNC32628.1 GGFJ01002457 MBW51598.1 KQ976418 KYM89482.1 ADTU01001095 ADTU01001096 CM000071 EAL25286.1 CH479181 EDW32210.1 CCAG010012977 GAMC01011724 JAB94831.1 CH916375 EDV98134.1 UFQS01000339 UFQT01000339 SSX02993.1 SSX23360.1 KQ414572 KOC71320.1 CVRI01000015 CRK89994.1 CH940648 EDW60508.1 OUUW01000001 SPP73609.1 GEDC01022843 GEDC01022098 JAS14455.1 JAS15200.1 GDHF01030967 JAI21347.1 CH902619 EDV37400.1 CH964154 EDW79724.2 AF135119 AE013599 BT031193 AAD31715.1 AAF57910.1 ABY20434.1 GBXI01011146 JAD03146.1 CM000362 CM002911 EDX07445.1 KMY94455.1 CM000158 EDW92196.1 KQ976948 KYN06925.1 MH366166 QBB01975.1 KQ982819 KYQ50205.1 CH480816 EDW48239.1 CH933808 EDW10650.1 NNAY01003485 OXU19347.1 DS235886 EEB20418.1 CP012524 ALC40525.1 CH954179 EDV55664.1 GAKP01021015 GAKP01021013 GAKP01021011 JAC37939.1 GDHF01018632 JAI33682.1 GBXI01005027 JAD09265.1 GL888217 EGI64795.1 KA649614 AFP64243.1 APGK01028062 APGK01028063 APGK01028064 APGK01028065 APGK01028066 APGK01028067 KB740684 ENN79594.1
OWR45527.1 KQ460297 KPJ16248.1 KZ149891 PZC79055.1 GANO01002591 JAB57280.1 KQ971338 EFA01753.1 APCN01002391 KK853014 KDR12501.1 ATLV01023973 KE525347 KFB50242.1 AAAB01008986 EAA00407.4 JXUM01023161 JXUM01023162 JXUM01023163 KQ560653 KXJ81428.1 NEVH01020337 PNF21664.1 GEZM01098931 JAV53636.1 AXCM01006417 DS232063 EDS33615.1 KZ288192 PBC34294.1 KQ434778 KZC04355.1 GFDL01006630 JAV28415.1 ADMH02000153 ETN67571.1 CH477271 EAT45270.1 JXUM01044332 JXUM01044333 KQ561396 KXJ78714.1 GALX01002028 JAB66438.1 GGFK01001193 MBW34514.1 GL444248 EFN61136.1 GGFM01000676 MBW21427.1 GGFK01001207 MBW34528.1 GGFK01001214 MBW34535.1 GGFJ01002281 MBW51422.1 GGFM01000740 MBW21491.1 GGFK01001455 MBW34776.1 AXCN02001106 BT126577 AEE61541.1 GAMD01001308 JAB00283.1 GL448228 EFN85065.1 KA649560 AFP64189.1 KK107488 QOIP01000012 EZA49891.1 RLU16301.1 KQ981855 KYN34836.1 JXJN01007649 JRES01000299 KNC32628.1 GGFJ01002457 MBW51598.1 KQ976418 KYM89482.1 ADTU01001095 ADTU01001096 CM000071 EAL25286.1 CH479181 EDW32210.1 CCAG010012977 GAMC01011724 JAB94831.1 CH916375 EDV98134.1 UFQS01000339 UFQT01000339 SSX02993.1 SSX23360.1 KQ414572 KOC71320.1 CVRI01000015 CRK89994.1 CH940648 EDW60508.1 OUUW01000001 SPP73609.1 GEDC01022843 GEDC01022098 JAS14455.1 JAS15200.1 GDHF01030967 JAI21347.1 CH902619 EDV37400.1 CH964154 EDW79724.2 AF135119 AE013599 BT031193 AAD31715.1 AAF57910.1 ABY20434.1 GBXI01011146 JAD03146.1 CM000362 CM002911 EDX07445.1 KMY94455.1 CM000158 EDW92196.1 KQ976948 KYN06925.1 MH366166 QBB01975.1 KQ982819 KYQ50205.1 CH480816 EDW48239.1 CH933808 EDW10650.1 NNAY01003485 OXU19347.1 DS235886 EEB20418.1 CP012524 ALC40525.1 CH954179 EDV55664.1 GAKP01021015 GAKP01021013 GAKP01021011 JAC37939.1 GDHF01018632 JAI33682.1 GBXI01005027 JAD09265.1 GL888217 EGI64795.1 KA649614 AFP64243.1 APGK01028062 APGK01028063 APGK01028064 APGK01028065 APGK01028066 APGK01028067 KB740684 ENN79594.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000076407
UP000007266
+ More
UP000075840 UP000075903 UP000027135 UP000075920 UP000030765 UP000075880 UP000007062 UP000069272 UP000069940 UP000249989 UP000235965 UP000075882 UP000075884 UP000075900 UP000075883 UP000002320 UP000075885 UP000242457 UP000076502 UP000000673 UP000005203 UP000075881 UP000008820 UP000000311 UP000192223 UP000075886 UP000075902 UP000008237 UP000095301 UP000078200 UP000053097 UP000279307 UP000092445 UP000095300 UP000092443 UP000078541 UP000092460 UP000037069 UP000078540 UP000005205 UP000001819 UP000008744 UP000091820 UP000092444 UP000001070 UP000053825 UP000183832 UP000192221 UP000008792 UP000268350 UP000007801 UP000007798 UP000000803 UP000000304 UP000002282 UP000078542 UP000075809 UP000001292 UP000009192 UP000215335 UP000009046 UP000002358 UP000092553 UP000008711 UP000076408 UP000007755 UP000019118
UP000075840 UP000075903 UP000027135 UP000075920 UP000030765 UP000075880 UP000007062 UP000069272 UP000069940 UP000249989 UP000235965 UP000075882 UP000075884 UP000075900 UP000075883 UP000002320 UP000075885 UP000242457 UP000076502 UP000000673 UP000005203 UP000075881 UP000008820 UP000000311 UP000192223 UP000075886 UP000075902 UP000008237 UP000095301 UP000078200 UP000053097 UP000279307 UP000092445 UP000095300 UP000092443 UP000078541 UP000092460 UP000037069 UP000078540 UP000005205 UP000001819 UP000008744 UP000091820 UP000092444 UP000001070 UP000053825 UP000183832 UP000192221 UP000008792 UP000268350 UP000007801 UP000007798 UP000000803 UP000000304 UP000002282 UP000078542 UP000075809 UP000001292 UP000009192 UP000215335 UP000009046 UP000002358 UP000092553 UP000008711 UP000076408 UP000007755 UP000019118
Pfam
Interpro
IPR002861
Reeler_dom
+ More
IPR038678 Spondin_N_sf
IPR002223 Kunitz_BPTI
IPR042307 Reeler_sf
IPR009465 Spondin_N
IPR036383 TSP1_rpt_sf
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR000884 TSP1_rpt
IPR019141 DUF2045
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR010920 LSM_dom_sf
IPR034109 Lsm11_M
IPR001163 LSM_dom_euk/arc
IPR038678 Spondin_N_sf
IPR002223 Kunitz_BPTI
IPR042307 Reeler_sf
IPR009465 Spondin_N
IPR036383 TSP1_rpt_sf
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR000884 TSP1_rpt
IPR019141 DUF2045
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR010920 LSM_dom_sf
IPR034109 Lsm11_M
IPR001163 LSM_dom_euk/arc
ProteinModelPortal
H9IV11
A0A2A4JL45
A0A2H1WUG2
A0A212EVP3
A0A194REI6
A0A2W1C015
+ More
U5ETX1 A0A182X6R7 D2A031 A0A182HH71 A0A182US71 A0A067QSB9 A0A182W1R7 A0A084WJ48 A0A182IWE1 Q7PZ75 A0A182FW84 A0A182H1I6 A0A2J7PZ95 A0A182L4P3 A0A182N3K1 A0A1Y1K2N4 A0A182RVE0 A0A182MDJ6 B0WRZ7 A0A182PSA0 A0A2A3ES07 A0A154NXF1 A0A1Q3FLK1 W5JTW2 A0A088AUJ4 A0A182KA48 Q17FH5 A0A182G628 V5H0T5 A0A2M4A123 A0A1S4F4R7 E2AZK3 A0A2M3YYU8 A0A2M4A1L1 A0A2M4A1B5 A0A2M4BEF7 A0A2M3YZ25 A0A2M4A1T8 A0A1W4XFH7 A0A182QMH4 A0A182TJA2 J3JTM0 T1EA54 E2BGT3 T1PNB9 A0A1I8MX43 A0A1A9UVC8 A0A026W2S5 A0A1B0ACU2 A0A1I8PHY5 A0A1A9Y5X7 A0A195F377 A0A1B0B2H8 A0A0L0CJP3 A0A2M4BFK2 A0A195BSB4 A0A158P1Q3 Q290S6 B4GBA8 A0A1A9WIB2 A0A1B0FAY9 W8BN93 B4JVZ6 A0A336KJF1 A0A0L7RKC0 A0A1J1HR27 A0A1W4VBY5 B4LJC8 A0A3B0JJI0 A0A1B6CP24 A0A0K8U3T0 B3MD67 B4N5T5 Q7KN04 A0A0A1WWX3 B4QIJ8 B4P8H9 A0A195D1Y4 A0A411G7Z9 A0A151WRA0 B4HMH1 B4KTS4 A0A232ELX5 E0W462 K7IZ87 A0A0M4ESX8 B3NP25 A0A034V5Z7 A0A182YL14 A0A0K8V430 A0A0A1XE95 F4WMF1 T1PLM0 N6TGR9
U5ETX1 A0A182X6R7 D2A031 A0A182HH71 A0A182US71 A0A067QSB9 A0A182W1R7 A0A084WJ48 A0A182IWE1 Q7PZ75 A0A182FW84 A0A182H1I6 A0A2J7PZ95 A0A182L4P3 A0A182N3K1 A0A1Y1K2N4 A0A182RVE0 A0A182MDJ6 B0WRZ7 A0A182PSA0 A0A2A3ES07 A0A154NXF1 A0A1Q3FLK1 W5JTW2 A0A088AUJ4 A0A182KA48 Q17FH5 A0A182G628 V5H0T5 A0A2M4A123 A0A1S4F4R7 E2AZK3 A0A2M3YYU8 A0A2M4A1L1 A0A2M4A1B5 A0A2M4BEF7 A0A2M3YZ25 A0A2M4A1T8 A0A1W4XFH7 A0A182QMH4 A0A182TJA2 J3JTM0 T1EA54 E2BGT3 T1PNB9 A0A1I8MX43 A0A1A9UVC8 A0A026W2S5 A0A1B0ACU2 A0A1I8PHY5 A0A1A9Y5X7 A0A195F377 A0A1B0B2H8 A0A0L0CJP3 A0A2M4BFK2 A0A195BSB4 A0A158P1Q3 Q290S6 B4GBA8 A0A1A9WIB2 A0A1B0FAY9 W8BN93 B4JVZ6 A0A336KJF1 A0A0L7RKC0 A0A1J1HR27 A0A1W4VBY5 B4LJC8 A0A3B0JJI0 A0A1B6CP24 A0A0K8U3T0 B3MD67 B4N5T5 Q7KN04 A0A0A1WWX3 B4QIJ8 B4P8H9 A0A195D1Y4 A0A411G7Z9 A0A151WRA0 B4HMH1 B4KTS4 A0A232ELX5 E0W462 K7IZ87 A0A0M4ESX8 B3NP25 A0A034V5Z7 A0A182YL14 A0A0K8V430 A0A0A1XE95 F4WMF1 T1PLM0 N6TGR9
PDB
3Q13
E-value=3.03052e-51,
Score=513
Ontologies
GO
Topology
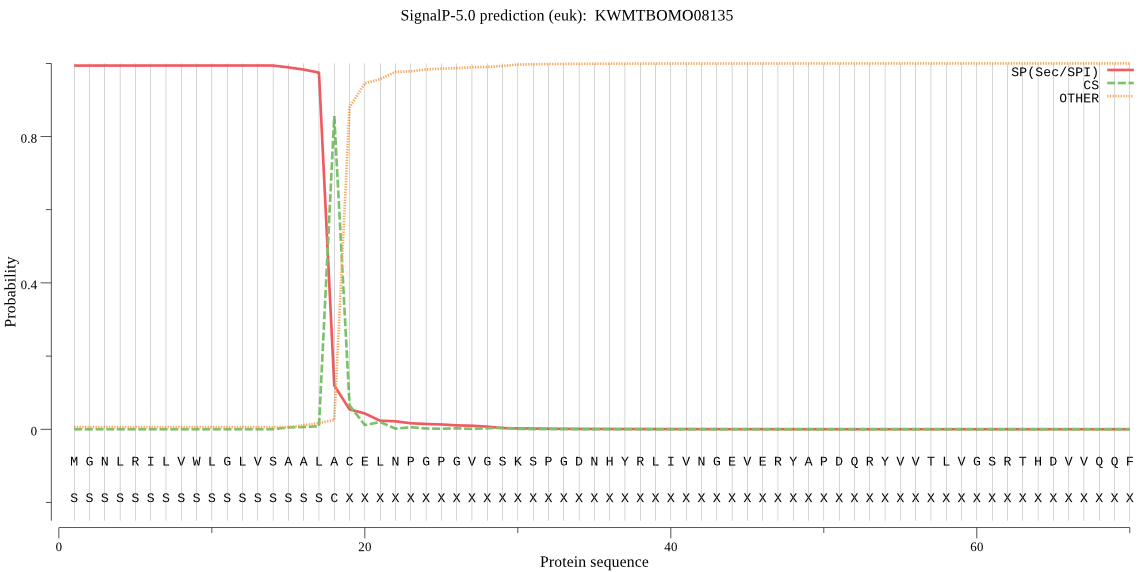
SignalP
Position: 1 - 18,
Likelihood: 0.993801
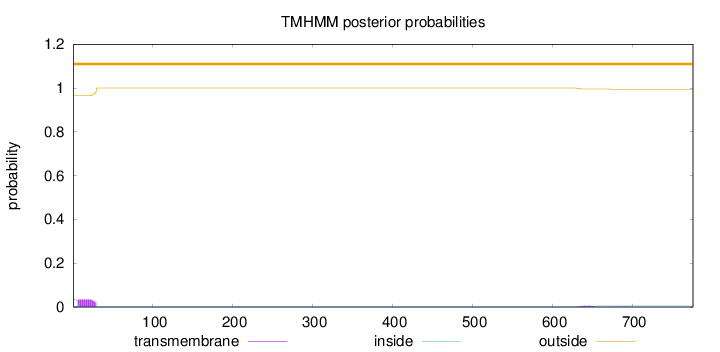
Length:
776
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.854489999999997
Exp number, first 60 AAs:
0.75024
Total prob of N-in:
0.03434
outside
1 - 776
Population Genetic Test Statistics
Pi
16.815065
Theta
18.210043
Tajima's D
-0.697362
CLR
1.044025
CSRT
0.199090045497725
Interpretation
Uncertain
