Gene
KWMTBOMO08134
Pre Gene Modal
BGIBMGA001074
Annotation
PREDICTED:_probable_phospholipid-transporting_ATPase_IA_isoform_X1_[Bombyx_mori]
Full name
Phospholipid-transporting ATPase
Location in the cell
PlasmaMembrane Reliability : 4.729
Sequence
CDS
ATGGGTCTAAACGATGTCGACATGGCGTTAGCTTTGAGAGAGTACGTGGTGAGCTTCTTGCAGGAGCACAACATTATGGCGCCTTTCTCCAGGTTCTATAGGGCCATGGTGCTCAAAATGAACCAGCATGAAGATGAAGCAACTACGTCAGGTATCACCGACGGTGCCACATCCGATCAGCAAAACCGTGTCATTTTCGTCAACAGACCGCAGCCCCAGAAGTTCGTCTCGAACAGGATCTCTACTGCTAAATACAGCGTGCCGTCCTTCGTTCCCTTATTCCTGTTCGAGCAGTTCCGTCGGTATTCCAACTGCTTCTTCCTGTTGATAGCCCTGCTGCAGCAGATCCCTGACGTGTCGCCGACGGGACGGTGGACAACCCTCACTCCTCTCATACTCATACTGAGCGTTAGCGCCATCAAGGAGATTGTTGAAGATTTTAAGCGGCACCGAGCGGACGACGAGACCAACCGCAGGCTGGTGGAGGTGCTGCGGGAGCGGCGCTGGGTCGCCATACGATGGGAGCATCTACAGATCGGCGACATCTGCAAAGTGCTCAACAACCAGTTCTTCCCAGCCGACCTGGTGCTGCTGGCCTCTAGCGAGCCCCAGGGCATATCCTTCATAGAGACCAGCAACCTGGACGGAGAGACCAACCTGAAGATCCGGCAGGCGCAGCCCGACACGGCCCGCCTGGACGCCGCCCCCGCCCTGGCGGACTTCCGCGCCACCGTGCAGTGCGAGCCGCCCAACCGACATCTATACGAATTCAATGGATTGCTAAAGGAGGCCAATGTCAAGACGCTGCCTCTGGGCTTGGACCAGATGCTCCTCCGGGGGGCGATGCTGCGGAACACGGCCTGGGTGCACGGCGTGGTGGTGTACACGGGACACGAAACTAAACTCATGAAGAATTCTACTAAAGCCCCGTTGAAGCGCTCATCGATCGACCGCCAGACCAACACCCACATCTTGATGCTGTTCATCATCCTCCTGGCGCTGTCGCTGCTCAGCGCCGGCTTCAACGAGTTCTGGATGCGGAACCACAATGACTGGTATATCGGCCTCGAAGAGGCGCAGAACGCGCACTTCGGGTTCAACTTCCTGACGTTCCTGATCCTGTACAACAACCTCATACCGATATCGCTGCAGGTCACCGCCGAGATAGTTCGCTTCTTCCAGGCCAAGTTCATCTCCATGGACAGCGAGATGTACGACCCGGCCAGCGACACGGCGGCCATGGCGCGGACCTCCAACCTCAACGAGGAGCTGGGCATGGTGCGCTACGTGTTCAGCGACAAGACCGGCACCCTCACGCAGAACGTCATGGTCTTCCACAAGTGCACCATCGCAGAGGTGATCTACAGCCGGCCGGGGCCCACGGAGCGGCTGGAGGACACGCCGCTGTACCAGAACCTGACGCGGGAGCACCCCACGGCGCCCGTCATCCGCGAGTTCCTCATCATGCTGGCCATCTGCCACACCGTCATCCCCGAGACCAAGGGGGACACCGTGGACTACCACGCCGCTTCCCCCGACGAGCGCGCGCTGGTGACGGGCGCCGCGGCCTTCGGCTTCTCGTTCTGCACGCGCACGCCCAGCCACGTGCACGTGCGCGCGCTGGGGGACACGCTGCGCTACCAGCTGCTGCACGTGCTCGACTTCACCTCCGCCAGGAAGCGCATGTCCGTCATCGTGCGCACGCCGGAAGGGACCATCAAGCTGTACTGCAAGGGCGCGGACAGCGTGATCTACAGCCGGCTGTCCGGCGGAGACAGCGCGCCCTTCGCGGCGGCCACCCTGGCGCACCTGGAGCACTTCGCCGCGGAGGGGCTCCGGACCCTCGTGTTCGCCGTCGCCGACATCCCCGAGCAGGTCTACCAGGACTGGTCCAACACCTACCACAAGGCGAGCATCGCGATACAGGACCGCGAGCAGAAGATCGAGGAAGCTGCCGATCTCGTAGAGAACAACCTGAGGCTGCTCGGAGCCACTGCCATTGAAGACAAGCTGCAGGACGGCGTGCCGGAGACCATCGCCGCGCTCCTGAAGGCCAACATCAACGTGTGGGTGCTCACGGGAGACAAGCAGGAGACCGCCATCAACATCGCGCACTCCGCGCGGCTCATCCACACCGCCATGCCGCTGCTCATACTCAACGAGGACAGCCTCGACGGCACCCGGGAGTCCATGTCGCGCCACGCCATAGACTTCGGAGACAACCTCCGGAAGCAGAACGACGTGGCGCTCGTCATCGACGGAAAGAGCCTCAAGTACGCGATGGGCTGCGACCTGAAGAAGGACTTCCTGGACCTGTGCATATCCTGCAAGGTGGTGGTGTGCTGCAGGGTGTCGCCCATACAGAAGGCCGAGGTGGTGGAGCTGGTGTCGGGCGCCACGGGGGCGGTGACGCTGGCCATCGGCGACGGAGCCAACGACGTGGCCATGATCCAGCGCGCGTCCGTCGGCGTCGGCATCTCCGGCGTCGAGGGGCTGCAGGCCGTCTGCGCCTCCGACTACAGCATCGCGCAGTTCCGTTTCCTGCTCCGCCTGCTGCTGGTGCACGGAGCCTGGAACTACTCGCGAATCAGCAAACTCATTCTGTATTCATTCTACAAAAACATCTGTCTGTACGTCATCGAACTGTGGTTCGCCATATACTCTGCTTGGTCGGGGCAGATCCTGTTCGAGCGGTGGACCATCGGCTTCTACAACGTTATCTTCACTGCGCTGCCACCCTTCGCCATCGGACTCTTCGACAAGCTGTGCTCACCGGAGATCATGCTCAGGCACCCCATCCTGTACATCCCGTCGCAACAAGGGCTGCTGTTCAACGTGCGCGTGTTCTGGGTGTGGGCGGTCAACGCGCTGCTGCACTCCGTGCTGCTGTTCTGGCTGCCCGTGGCGCTCGCCGACCACCACGTGCTGTGGTCCTCGGGCAAGGACGGCGGCTACCTGGTGCTGGGGAACCTGGTCTACACCTACGTGGTTCTGACCGTGTGTCTCAAAGCCGGGCTGGCCACGCACTCGTGGACGTGGGTCACGCACGTCTCCATATGGGGGTCCTTAGCTCTTTGGCTGCTATTCTTACTTATATACAGTAACATATACCCAACGATTCTGATCGGAGCCGTGATGCTGGGCATGGACCGGATGGTGTTCGGCTCCCTGGTCTTCTGGTTCGGCCTCATCCTCATCCCGATAGCGACCCTCATACCGGACCTCGTCGTCACCGTCGTGCGCAACTCCGCCTTCAAATCCGCCACCGAGGCCGTGCGGGAGAGCGAGCTGAAGCAGCGCGCGCCGCCCGCCCTGCTGGCCCAGCCGCGACACTCCCTCACGGAGACGGCGCGGCTGCTGCAGAACGTTCGCAGCGTGTTCACTCGGCGCTCCACCTCGCGGGCCGCCGTCGAGCTAGAGCTCTCGCATGGATTCGCTTTCTCGCAAGAGGAGGGCGGGAGTGTGTCCCAGGCGGACGTGGTCCGAGTGTACGACACAAGGCAGCCGAGACCGGAGCACTCTTGA
Protein
MGLNDVDMALALREYVVSFLQEHNIMAPFSRFYRAMVLKMNQHEDEATTSGITDGATSDQQNRVIFVNRPQPQKFVSNRISTAKYSVPSFVPLFLFEQFRRYSNCFFLLIALLQQIPDVSPTGRWTTLTPLILILSVSAIKEIVEDFKRHRADDETNRRLVEVLRERRWVAIRWEHLQIGDICKVLNNQFFPADLVLLASSEPQGISFIETSNLDGETNLKIRQAQPDTARLDAAPALADFRATVQCEPPNRHLYEFNGLLKEANVKTLPLGLDQMLLRGAMLRNTAWVHGVVVYTGHETKLMKNSTKAPLKRSSIDRQTNTHILMLFIILLALSLLSAGFNEFWMRNHNDWYIGLEEAQNAHFGFNFLTFLILYNNLIPISLQVTAEIVRFFQAKFISMDSEMYDPASDTAAMARTSNLNEELGMVRYVFSDKTGTLTQNVMVFHKCTIAEVIYSRPGPTERLEDTPLYQNLTREHPTAPVIREFLIMLAICHTVIPETKGDTVDYHAASPDERALVTGAAAFGFSFCTRTPSHVHVRALGDTLRYQLLHVLDFTSARKRMSVIVRTPEGTIKLYCKGADSVIYSRLSGGDSAPFAAATLAHLEHFAAEGLRTLVFAVADIPEQVYQDWSNTYHKASIAIQDREQKIEEAADLVENNLRLLGATAIEDKLQDGVPETIAALLKANINVWVLTGDKQETAINIAHSARLIHTAMPLLILNEDSLDGTRESMSRHAIDFGDNLRKQNDVALVIDGKSLKYAMGCDLKKDFLDLCISCKVVVCCRVSPIQKAEVVELVSGATGAVTLAIGDGANDVAMIQRASVGVGISGVEGLQAVCASDYSIAQFRFLLRLLLVHGAWNYSRISKLILYSFYKNICLYVIELWFAIYSAWSGQILFERWTIGFYNVIFTALPPFAIGLFDKLCSPEIMLRHPILYIPSQQGLLFNVRVFWVWAVNALLHSVLLFWLPVALADHHVLWSSGKDGGYLVLGNLVYTYVVLTVCLKAGLATHSWTWVTHVSIWGSLALWLLFLLIYSNIYPTILIGAVMLGMDRMVFGSLVFWFGLILIPIATLIPDLVVTVVRNSAFKSATEAVRESELKQRAPPALLAQPRHSLTETARLLQNVRSVFTRRSTSRAAVELELSHGFAFSQEEGGSVSQADVVRVYDTRQPRPEHS
Summary
Catalytic Activity
ATP + H(2)O + phospholipid(Side 1) = ADP + phosphate + phospholipid(Side 2).
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IV subfamily.
Feature
chain Phospholipid-transporting ATPase
Uniprot
H9IUZ4
A0A194RG10
A0A212EVK6
A0A1L8DEL1
U5EWK7
A0A1Q3G3M2
+ More
A0A1Q3G3L2 A0A067RBF1 A0A0L0BV56 A0A1L8DE92 A0A1I8P2W8 A0A1I8P2Y5 A0A1B0A519 A0A1I8NB71 A0A1I8NB83 Q17N94 A0A1A9W039 A0A336MCV4 A0A1A9XGU1 A0A336MA04 A0A1B0FNL8 A0A1B0BJF9 A0A1A9V9K6 N6W4Q3 A0A3B0JEZ8 B4KTN7 W8BBM3 W8ARP7 A0A0K8V6W8 A0A0K8VP29 A0A034V3T4 D2A294 A0A0Q5VZ68 A0A0Q9X8V3 A0A0P8YDR8 A0A1Y9IYG1 A0A0Q5VP98 A0A0Q5W0M8 A0A1W4VGI2 A0A0P8Y6Q7 A0A0R1DPX3 A0A1W4VH17 A0A182XC77 A0A0Q5VWW1 A0A1W4VGH7 A0A0Q5VMY5 A0A0R1DQ69 A0A182YK07 Q8T0I4 A0A0J9U0W3 A0A0R1DQU3 A0A1Y1MJW6 A0A0J9RBT3 A0A1Y1MD21 A0A0Q9W2Z2 A0A3L8D697 B7YZF5 A0A026WAN0 A0A0J9RCZ3 B7YZF6 A0A0R3NPB7 A0A151J935 A0A336MAX9 B4MIW7 A0A336ME27 E2A435 A0A195CI92 A0A1W4XFT1 A0A0K8VME7 A0A0K8UV14 A0A1Q3G3K0 A0A195B6B1 A0A0M9A0V8 A0A2M3ZFX1 A0A1Q3G3K8 V9I7B1 F4WL37 V9I9H6 A0A139WIQ2 A0A088AVA3 A0A151WM27 A0A2A3ET72 A0A310SAH7 A0A2M4BBB1 A0A0Q9X8Q2 U4UI06 A0A2M4BB13 A0A2M4CUY1 A0A182FKQ4 A0A0Q5W1W9 Q17N93 A0A2M4A747 A0A154P6V4 A0A0C9QBV8 A0A0Q5VX56 A0A1W4VH26 A0A1Y1MG03 A0A0R1DQ74 A0A1W4V4P7 D0Z766
A0A1Q3G3L2 A0A067RBF1 A0A0L0BV56 A0A1L8DE92 A0A1I8P2W8 A0A1I8P2Y5 A0A1B0A519 A0A1I8NB71 A0A1I8NB83 Q17N94 A0A1A9W039 A0A336MCV4 A0A1A9XGU1 A0A336MA04 A0A1B0FNL8 A0A1B0BJF9 A0A1A9V9K6 N6W4Q3 A0A3B0JEZ8 B4KTN7 W8BBM3 W8ARP7 A0A0K8V6W8 A0A0K8VP29 A0A034V3T4 D2A294 A0A0Q5VZ68 A0A0Q9X8V3 A0A0P8YDR8 A0A1Y9IYG1 A0A0Q5VP98 A0A0Q5W0M8 A0A1W4VGI2 A0A0P8Y6Q7 A0A0R1DPX3 A0A1W4VH17 A0A182XC77 A0A0Q5VWW1 A0A1W4VGH7 A0A0Q5VMY5 A0A0R1DQ69 A0A182YK07 Q8T0I4 A0A0J9U0W3 A0A0R1DQU3 A0A1Y1MJW6 A0A0J9RBT3 A0A1Y1MD21 A0A0Q9W2Z2 A0A3L8D697 B7YZF5 A0A026WAN0 A0A0J9RCZ3 B7YZF6 A0A0R3NPB7 A0A151J935 A0A336MAX9 B4MIW7 A0A336ME27 E2A435 A0A195CI92 A0A1W4XFT1 A0A0K8VME7 A0A0K8UV14 A0A1Q3G3K0 A0A195B6B1 A0A0M9A0V8 A0A2M3ZFX1 A0A1Q3G3K8 V9I7B1 F4WL37 V9I9H6 A0A139WIQ2 A0A088AVA3 A0A151WM27 A0A2A3ET72 A0A310SAH7 A0A2M4BBB1 A0A0Q9X8Q2 U4UI06 A0A2M4BB13 A0A2M4CUY1 A0A182FKQ4 A0A0Q5W1W9 Q17N93 A0A2M4A747 A0A154P6V4 A0A0C9QBV8 A0A0Q5VX56 A0A1W4VH26 A0A1Y1MG03 A0A0R1DQ74 A0A1W4V4P7 D0Z766
EC Number
7.6.2.1
Pubmed
EMBL
BABH01000052
BABH01000053
BABH01000054
KQ460297
KPJ16245.1
AGBW02012175
+ More
OWR45528.1 GFDF01009182 JAV04902.1 GANO01001371 JAB58500.1 GFDL01000645 JAV34400.1 GFDL01000655 JAV34390.1 KK852570 KDR21171.1 JRES01001289 KNC23912.1 GFDF01009379 JAV04705.1 CH477201 EAT48160.1 UFQT01000781 SSX27181.1 SSX27182.1 CCAG010015909 JXJN01015380 CM000071 ENO01787.1 OUUW01000001 SPP73870.1 CH933808 EDW09620.2 GAMC01019295 GAMC01019291 JAB87264.1 GAMC01019293 GAMC01019292 JAB87263.1 GDHF01017999 JAI34315.1 GDHF01011677 JAI40637.1 GAKP01022156 GAKP01022155 JAC36797.1 KQ971338 EFA02157.2 CH954179 KQS62750.1 KRG04764.1 CH902619 KPU77080.1 KQS62748.1 KQS62746.1 KPU77083.1 CM000158 KRJ99335.1 KQS62745.1 KQS62743.1 KRJ99336.1 AY069236 AE013599 AAL39381.1 AAM68573.1 CM002911 KMY93480.1 KRJ99342.1 GEZM01034879 JAV83567.1 KMY93482.1 GEZM01034880 GEZM01034874 JAV83573.1 CH940648 KRF79266.1 QOIP01000012 RLU15759.1 ACL83103.1 KK107353 EZA52084.1 KMY93484.1 ACL83109.1 KRT02810.1 KQ979471 KYN21420.1 SSX27180.1 CH963719 EDW72056.2 SSX27179.1 GL436526 EFN71801.1 KQ977720 KYN00428.1 GDHF01012252 JAI40062.1 GDHF01021961 JAI30353.1 GFDL01000662 JAV34383.1 KQ976587 KYM79719.1 KQ435794 KOX73593.1 GGFM01006716 MBW27467.1 GFDL01000661 JAV34384.1 JR036186 AEY56983.1 GL888207 EGI65000.1 JR036187 AEY56984.1 KYB27799.1 KQ982944 KYQ48851.1 KZ288192 PBC34339.1 KQ766603 OAD53651.1 GGFJ01001182 MBW50323.1 KRG04767.1 KB632308 ERL91978.1 GGFJ01001085 MBW50226.1 GGFL01004978 MBW69156.1 KQS62751.1 EAT48161.1 GGFK01003305 MBW36626.1 KQ434827 KZC07665.1 GBYB01011958 JAG81725.1 KQS62747.1 GEZM01034876 JAV83570.1 KRJ99340.1 BT100313 ACZ52625.1
OWR45528.1 GFDF01009182 JAV04902.1 GANO01001371 JAB58500.1 GFDL01000645 JAV34400.1 GFDL01000655 JAV34390.1 KK852570 KDR21171.1 JRES01001289 KNC23912.1 GFDF01009379 JAV04705.1 CH477201 EAT48160.1 UFQT01000781 SSX27181.1 SSX27182.1 CCAG010015909 JXJN01015380 CM000071 ENO01787.1 OUUW01000001 SPP73870.1 CH933808 EDW09620.2 GAMC01019295 GAMC01019291 JAB87264.1 GAMC01019293 GAMC01019292 JAB87263.1 GDHF01017999 JAI34315.1 GDHF01011677 JAI40637.1 GAKP01022156 GAKP01022155 JAC36797.1 KQ971338 EFA02157.2 CH954179 KQS62750.1 KRG04764.1 CH902619 KPU77080.1 KQS62748.1 KQS62746.1 KPU77083.1 CM000158 KRJ99335.1 KQS62745.1 KQS62743.1 KRJ99336.1 AY069236 AE013599 AAL39381.1 AAM68573.1 CM002911 KMY93480.1 KRJ99342.1 GEZM01034879 JAV83567.1 KMY93482.1 GEZM01034880 GEZM01034874 JAV83573.1 CH940648 KRF79266.1 QOIP01000012 RLU15759.1 ACL83103.1 KK107353 EZA52084.1 KMY93484.1 ACL83109.1 KRT02810.1 KQ979471 KYN21420.1 SSX27180.1 CH963719 EDW72056.2 SSX27179.1 GL436526 EFN71801.1 KQ977720 KYN00428.1 GDHF01012252 JAI40062.1 GDHF01021961 JAI30353.1 GFDL01000662 JAV34383.1 KQ976587 KYM79719.1 KQ435794 KOX73593.1 GGFM01006716 MBW27467.1 GFDL01000661 JAV34384.1 JR036186 AEY56983.1 GL888207 EGI65000.1 JR036187 AEY56984.1 KYB27799.1 KQ982944 KYQ48851.1 KZ288192 PBC34339.1 KQ766603 OAD53651.1 GGFJ01001182 MBW50323.1 KRG04767.1 KB632308 ERL91978.1 GGFJ01001085 MBW50226.1 GGFL01004978 MBW69156.1 KQS62751.1 EAT48161.1 GGFK01003305 MBW36626.1 KQ434827 KZC07665.1 GBYB01011958 JAG81725.1 KQS62747.1 GEZM01034876 JAV83570.1 KRJ99340.1 BT100313 ACZ52625.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000027135
UP000037069
UP000095300
+ More
UP000092445 UP000095301 UP000008820 UP000091820 UP000092443 UP000092444 UP000092460 UP000078200 UP000001819 UP000268350 UP000009192 UP000007266 UP000008711 UP000007801 UP000076407 UP000192221 UP000002282 UP000076408 UP000000803 UP000008792 UP000279307 UP000053097 UP000078492 UP000007798 UP000000311 UP000078542 UP000192223 UP000078540 UP000053105 UP000007755 UP000005203 UP000075809 UP000242457 UP000030742 UP000069272 UP000076502
UP000092445 UP000095301 UP000008820 UP000091820 UP000092443 UP000092444 UP000092460 UP000078200 UP000001819 UP000268350 UP000009192 UP000007266 UP000008711 UP000007801 UP000076407 UP000192221 UP000002282 UP000076408 UP000000803 UP000008792 UP000279307 UP000053097 UP000078492 UP000007798 UP000000311 UP000078542 UP000192223 UP000078540 UP000053105 UP000007755 UP000005203 UP000075809 UP000242457 UP000030742 UP000069272 UP000076502
Interpro
Gene 3D
ProteinModelPortal
H9IUZ4
A0A194RG10
A0A212EVK6
A0A1L8DEL1
U5EWK7
A0A1Q3G3M2
+ More
A0A1Q3G3L2 A0A067RBF1 A0A0L0BV56 A0A1L8DE92 A0A1I8P2W8 A0A1I8P2Y5 A0A1B0A519 A0A1I8NB71 A0A1I8NB83 Q17N94 A0A1A9W039 A0A336MCV4 A0A1A9XGU1 A0A336MA04 A0A1B0FNL8 A0A1B0BJF9 A0A1A9V9K6 N6W4Q3 A0A3B0JEZ8 B4KTN7 W8BBM3 W8ARP7 A0A0K8V6W8 A0A0K8VP29 A0A034V3T4 D2A294 A0A0Q5VZ68 A0A0Q9X8V3 A0A0P8YDR8 A0A1Y9IYG1 A0A0Q5VP98 A0A0Q5W0M8 A0A1W4VGI2 A0A0P8Y6Q7 A0A0R1DPX3 A0A1W4VH17 A0A182XC77 A0A0Q5VWW1 A0A1W4VGH7 A0A0Q5VMY5 A0A0R1DQ69 A0A182YK07 Q8T0I4 A0A0J9U0W3 A0A0R1DQU3 A0A1Y1MJW6 A0A0J9RBT3 A0A1Y1MD21 A0A0Q9W2Z2 A0A3L8D697 B7YZF5 A0A026WAN0 A0A0J9RCZ3 B7YZF6 A0A0R3NPB7 A0A151J935 A0A336MAX9 B4MIW7 A0A336ME27 E2A435 A0A195CI92 A0A1W4XFT1 A0A0K8VME7 A0A0K8UV14 A0A1Q3G3K0 A0A195B6B1 A0A0M9A0V8 A0A2M3ZFX1 A0A1Q3G3K8 V9I7B1 F4WL37 V9I9H6 A0A139WIQ2 A0A088AVA3 A0A151WM27 A0A2A3ET72 A0A310SAH7 A0A2M4BBB1 A0A0Q9X8Q2 U4UI06 A0A2M4BB13 A0A2M4CUY1 A0A182FKQ4 A0A0Q5W1W9 Q17N93 A0A2M4A747 A0A154P6V4 A0A0C9QBV8 A0A0Q5VX56 A0A1W4VH26 A0A1Y1MG03 A0A0R1DQ74 A0A1W4V4P7 D0Z766
A0A1Q3G3L2 A0A067RBF1 A0A0L0BV56 A0A1L8DE92 A0A1I8P2W8 A0A1I8P2Y5 A0A1B0A519 A0A1I8NB71 A0A1I8NB83 Q17N94 A0A1A9W039 A0A336MCV4 A0A1A9XGU1 A0A336MA04 A0A1B0FNL8 A0A1B0BJF9 A0A1A9V9K6 N6W4Q3 A0A3B0JEZ8 B4KTN7 W8BBM3 W8ARP7 A0A0K8V6W8 A0A0K8VP29 A0A034V3T4 D2A294 A0A0Q5VZ68 A0A0Q9X8V3 A0A0P8YDR8 A0A1Y9IYG1 A0A0Q5VP98 A0A0Q5W0M8 A0A1W4VGI2 A0A0P8Y6Q7 A0A0R1DPX3 A0A1W4VH17 A0A182XC77 A0A0Q5VWW1 A0A1W4VGH7 A0A0Q5VMY5 A0A0R1DQ69 A0A182YK07 Q8T0I4 A0A0J9U0W3 A0A0R1DQU3 A0A1Y1MJW6 A0A0J9RBT3 A0A1Y1MD21 A0A0Q9W2Z2 A0A3L8D697 B7YZF5 A0A026WAN0 A0A0J9RCZ3 B7YZF6 A0A0R3NPB7 A0A151J935 A0A336MAX9 B4MIW7 A0A336ME27 E2A435 A0A195CI92 A0A1W4XFT1 A0A0K8VME7 A0A0K8UV14 A0A1Q3G3K0 A0A195B6B1 A0A0M9A0V8 A0A2M3ZFX1 A0A1Q3G3K8 V9I7B1 F4WL37 V9I9H6 A0A139WIQ2 A0A088AVA3 A0A151WM27 A0A2A3ET72 A0A310SAH7 A0A2M4BBB1 A0A0Q9X8Q2 U4UI06 A0A2M4BB13 A0A2M4CUY1 A0A182FKQ4 A0A0Q5W1W9 Q17N93 A0A2M4A747 A0A154P6V4 A0A0C9QBV8 A0A0Q5VX56 A0A1W4VH26 A0A1Y1MG03 A0A0R1DQ74 A0A1W4V4P7 D0Z766
PDB
6A69
E-value=2.6922e-14,
Score=196
Ontologies
GO
PANTHER
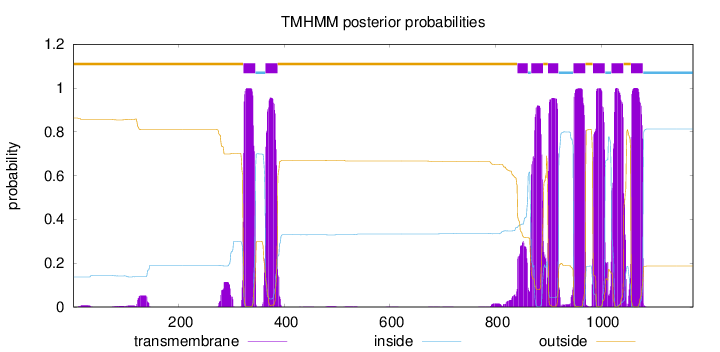
Topology
Subcellular location
Membrane
Length:
1174
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
185.15398
Exp number, first 60 AAs:
0.14565
Total prob of N-in:
0.13682
outside
1 - 322
TMhelix
323 - 345
inside
346 - 364
TMhelix
365 - 387
outside
388 - 841
TMhelix
842 - 861
inside
862 - 867
TMhelix
868 - 890
outside
891 - 899
TMhelix
900 - 919
inside
920 - 947
TMhelix
948 - 970
outside
971 - 984
TMhelix
985 - 1007
inside
1008 - 1019
TMhelix
1020 - 1042
outside
1043 - 1056
TMhelix
1057 - 1079
inside
1080 - 1174
Population Genetic Test Statistics
Pi
15.938211
Theta
17.907273
Tajima's D
-0.853523
CLR
1.029669
CSRT
0.16289185540723
Interpretation
Uncertain
