Pre Gene Modal
BGIBMGA001073
Annotation
PREDICTED:_cholinephosphotransferase_1_isoform_X3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.889
Sequence
CDS
ATGCAGTTCTACAAAGAAAGAATACTCAATGCTGCGCAGTTGAAAAGACTAAGTGAACACAAATACTCTTGCACCAGTGCCAGCGTTCTTGACGCATGGCTCCAGCCGTGGTGGTGTTGGCTGGTATCAAAGACGCCTTTATGGCTAGCTCCGAATCTCATCACAATTTTAGGTCTAATTGTAAATATAGTAACCACGCTTATATTAGTATGGTACAGTCCTGATGCCAGACAAGACCCACCAAGATGGGCATGTGCTCTGTGCGCCTTAGGTGTCTTTGTTTATCAGAGCCTGGATGCCATTGATGGTAAACAAGCAAGACGGACAGGCAGTCAATCTCCTTTGGGAGAATTGTTTGATCATGGATGTGACAGCATATCCACTGTGTTCATAGCTTTGGGTGCTTGTATTGCTGTCAAGCTCGGAGAGTATCCAACCTGGATGTTCTTTCAGTGTTTCTGTGCGATGACTCTGTTCTACTGCGCACACTGGCAAGCCTACGTAACGGGGACGTTGAAGATGGGTCGGATAGACGTGACCGAGGCTCAGTTCTCGATTATCGGCATACATCTAATATCCGCTGTCCTCGGACCCGACTTCTGGTCCACTCAGGTCGGTGGTGGACTCGAAATCCGCTACTCGGTGGCTCTGGCCGCTATCGTTGGAGCCTTCCTCACCATAGGGACGCTGGCTGCCGCTATCGTCAGCGGCGGCGTTGGGAAAAACGGCTCCACCGTCGCTGGCACGAGCATCCTGTCTCCGGTGATACCGTTCTCGCTGGTCGTGGTGCCGGCGTTTATCATCTTCCAGAAGAGCGAGTCGCACGTGTACGAGAACCACCCGGCGCTCTACATCTTAGCGTTCGGCATGGTCACGGCGAAGGTCACGAACAAACTGGTGGTGGCTCACATGACTAAAAGCGAGATGGAGTACTACGACTGGTCACTGCTGGGGCCGGCGATGCTGTTCCTTAACCAGTACTTCAACAACGCGCTGCCGGAGTACTGGGTGCTCTGGCTCTGCACGGTGTGGGTGTGCGGGGAGCTGGTGCGCTACTGCGGACAGGTCTGCCGCGAGATCTGCGACCACCTCGACATCCAGCTGTTCCGCATCCCGCGCGCCGCGCCCGCCCCCGCCTCCGCCTCCGCGCCCGCGCACAGCAAGGCCGCGCCCGACAGAAACGGTACGCGACGCCACGCCGCCAAGCACGCGAGACGACAGCCAGCCAACTAA
Protein
MQFYKERILNAAQLKRLSEHKYSCTSASVLDAWLQPWWCWLVSKTPLWLAPNLITILGLIVNIVTTLILVWYSPDARQDPPRWACALCALGVFVYQSLDAIDGKQARRTGSQSPLGELFDHGCDSISTVFIALGACIAVKLGEYPTWMFFQCFCAMTLFYCAHWQAYVTGTLKMGRIDVTEAQFSIIGIHLISAVLGPDFWSTQVGGGLEIRYSVALAAIVGAFLTIGTLAAAIVSGGVGKNGSTVAGTSILSPVIPFSLVVVPAFIIFQKSESHVYENHPALYILAFGMVTAKVTNKLVVAHMTKSEMEYYDWSLLGPAMLFLNQYFNNALPEYWVLWLCTVWVCGELVRYCGQVCREICDHLDIQLFRIPRAAPAPASASAPAHSKAAPDRNGTRRHAAKHARRQPAN
Summary
Similarity
Belongs to the CDP-alcohol phosphatidyltransferase class-I family.
Uniprot
H9IUZ3
A0A2A4J7T5
A0A2A4J6I1
A0A2A4J7J8
A0A3S2LSD2
A0A026WR91
+ More
A0A2J7PMM8 A0A067RQ11 K7J4I6 A0A1Q3FTS1 A0A1Q3FTT4 A0A1Q3FTQ2 A0A1Q3FX27 A0A1Q3FU90 U5ESN6 D2A0U9 A0A1Q3FTQ8 A0A1Q3FY07 A0A1Q3FXE0 V9I944 A0A1Q3G2T6 A0A1Q3FV56 A0A1Q3FUW0 A0A1Q3G397 A0A1Q3G329 A0A1Q3FXP1 A0A1Q3FUT5 A0A1Q3FUQ2 A0A1Q3FXX3 A0A1Q3G3Q2 Q5TMM4 Q16GF6 A0A1Y1MJH0 V9IBC0 A0A1Y1MJD2 A0A023EU02 A0A1W4X2U1 A0A1B6GT41 A0A1B6IUG1 A0A2R7VVK9 A0A1Y1ML92 A0A3B0IZQ9 A0A1W4X1Q6 A0A1Y1MMA7 T1PFL7 A0A0Q9WLY1 A0A1I8PM53 A0A0R3NN33 B4KML0 A0A1W4WS27 B3MGZ2 A0A1W4V4E3 D3TN27 B3NRK4 A0A2M4AB71 E0VXP8 A0A0P4VN76 A0A0A1X798 A0A1B6L9K1 A0A1B6LXK0 R4FMY2 B4P4H0 A0A0K8VNW2 A0A034VXL9 A0A0J9U111 A0A1B6G466 A0A0K8TT93 A0A3B0JKI2 Q28ZT2 A1Z9E0 U3GRY8 A0A1I8PM90 A0A1I8N2M4 B4MCL8 A0A0A9Z421 A0A0N8P0H0 A0A069DT05 A0A0A9W9M4 A0A1B6GJY7 A0A3B0IZG4 A0A0R3NN50 A0A0V0G3U8 A0A1B6EL77 A0A1I8N2N6 A0A1W4VGM2 A0A0Q5VX37 A0A0K8WK95 A0A1I8PM80 A0A034W061 W8AP52 A0A0A1XQY5 A0A0R1DQ53 A0A1L8DYA7 A0A0J9RBY5 A0A1B6KY80 A0A034W2R5 A0A1I8N2R1 A1Z9D9 A0A0P8ZRN5 A0A0K8W8Z9
A0A2J7PMM8 A0A067RQ11 K7J4I6 A0A1Q3FTS1 A0A1Q3FTT4 A0A1Q3FTQ2 A0A1Q3FX27 A0A1Q3FU90 U5ESN6 D2A0U9 A0A1Q3FTQ8 A0A1Q3FY07 A0A1Q3FXE0 V9I944 A0A1Q3G2T6 A0A1Q3FV56 A0A1Q3FUW0 A0A1Q3G397 A0A1Q3G329 A0A1Q3FXP1 A0A1Q3FUT5 A0A1Q3FUQ2 A0A1Q3FXX3 A0A1Q3G3Q2 Q5TMM4 Q16GF6 A0A1Y1MJH0 V9IBC0 A0A1Y1MJD2 A0A023EU02 A0A1W4X2U1 A0A1B6GT41 A0A1B6IUG1 A0A2R7VVK9 A0A1Y1ML92 A0A3B0IZQ9 A0A1W4X1Q6 A0A1Y1MMA7 T1PFL7 A0A0Q9WLY1 A0A1I8PM53 A0A0R3NN33 B4KML0 A0A1W4WS27 B3MGZ2 A0A1W4V4E3 D3TN27 B3NRK4 A0A2M4AB71 E0VXP8 A0A0P4VN76 A0A0A1X798 A0A1B6L9K1 A0A1B6LXK0 R4FMY2 B4P4H0 A0A0K8VNW2 A0A034VXL9 A0A0J9U111 A0A1B6G466 A0A0K8TT93 A0A3B0JKI2 Q28ZT2 A1Z9E0 U3GRY8 A0A1I8PM90 A0A1I8N2M4 B4MCL8 A0A0A9Z421 A0A0N8P0H0 A0A069DT05 A0A0A9W9M4 A0A1B6GJY7 A0A3B0IZG4 A0A0R3NN50 A0A0V0G3U8 A0A1B6EL77 A0A1I8N2N6 A0A1W4VGM2 A0A0Q5VX37 A0A0K8WK95 A0A1I8PM80 A0A034W061 W8AP52 A0A0A1XQY5 A0A0R1DQ53 A0A1L8DYA7 A0A0J9RBY5 A0A1B6KY80 A0A034W2R5 A0A1I8N2R1 A1Z9D9 A0A0P8ZRN5 A0A0K8W8Z9
Pubmed
19121390
24508170
24845553
20075255
18362917
19820115
+ More
12364791 17510324 28004739 24945155 25315136 17994087 15632085 20353571 18057021 20566863 27129103 25830018 17550304 25348373 22936249 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 26334808 24495485
12364791 17510324 28004739 24945155 25315136 17994087 15632085 20353571 18057021 20566863 27129103 25830018 17550304 25348373 22936249 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 26334808 24495485
EMBL
BABH01000055
BABH01000056
NWSH01002662
PCG67728.1
PCG67727.1
PCG67726.1
+ More
RSAL01000294 RVE42849.1 KK107119 EZA58555.1 NEVH01023972 PNF17590.1 KK852498 KDR22680.1 AAZX01002341 AAZX01009423 AAZX01019112 GFDL01004078 JAV30967.1 GFDL01004070 JAV30975.1 GFDL01004071 JAV30974.1 GFDL01002874 JAV32171.1 GFDL01004052 JAV30993.1 GANO01003184 JAB56687.1 KQ971338 EFA02564.1 GFDL01004056 JAV30989.1 GFDL01002683 JAV32362.1 GFDL01002788 JAV32257.1 JR037114 AEY57590.1 GFDL01000910 JAV34135.1 GFDL01003732 JAV31313.1 GFDL01003710 JAV31335.1 GFDL01000771 JAV34274.1 GFDL01000833 JAV34212.1 GFDL01002803 JAV32242.1 GFDL01003718 JAV31327.1 GFDL01003714 JAV31331.1 GFDL01002628 JAV32417.1 GFDL01000656 JAV34389.1 AAAB01008986 EAL38873.2 CH478283 EAT33324.1 GEZM01030030 JAV85833.1 JR037113 AEY57589.1 GEZM01030028 JAV85834.1 GAPW01001217 JAC12381.1 GECZ01004170 JAS65599.1 GECU01017164 JAS90542.1 KK854114 PTY11524.1 GEZM01030035 JAV85828.1 OUUW01000001 SPP73755.1 GEZM01030033 JAV85830.1 KA646698 AFP61327.1 CH940659 KRF85706.1 CM000071 KRT02346.1 CH933808 EDW10857.2 CH902619 EDV37910.2 EZ422829 ADD19105.1 CH954179 EDV56156.2 KQS62729.1 GGFK01004714 MBW38035.1 DS235833 EEB18154.1 GDKW01002240 JAI54355.1 GBXI01007290 JAD07002.1 GEBQ01023761 GEBQ01021301 GEBQ01019585 JAT16216.1 JAT18676.1 JAT20392.1 GEBQ01011538 JAT28439.1 ACPB03023223 GAHY01001559 JAA75951.1 CM000158 EDW90609.2 GDHF01031185 GDHF01015416 GDHF01011730 JAI21129.1 JAI36898.1 JAI40584.1 GAKP01010888 GAKP01010886 JAC48066.1 CM002911 KMY93515.1 GECZ01012534 JAS57235.1 GDAI01000230 JAI17373.1 SPP73756.1 EAL25531.3 AE013599 AAF58369.1 BT150228 AGU12789.1 EDW71406.2 GBHO01007049 JAG36555.1 KPU77100.1 GBGD01001661 JAC87228.1 GBHO01039100 GBRD01000766 JAG04504.1 JAG65055.1 GECZ01007069 JAS62700.1 SPP73754.1 KRT02345.1 GECL01003409 JAP02715.1 GECZ01031118 JAS38651.1 KQS62731.1 GDHF01010456 GDHF01000743 JAI41858.1 JAI51571.1 GAKP01010887 GAKP01010883 JAC48069.1 GAMC01018758 JAB87797.1 GBXI01001304 GBXI01000381 JAD12988.1 JAD13911.1 KRJ99320.1 GFDF01002653 JAV11431.1 KMY93513.1 GEBQ01023569 JAT16408.1 GAKP01010885 GAKP01010884 JAC48068.1 AAF58370.1 KPU77101.1 GDHF01012387 GDHF01004636 GDHF01002563 JAI39927.1 JAI47678.1 JAI49751.1
RSAL01000294 RVE42849.1 KK107119 EZA58555.1 NEVH01023972 PNF17590.1 KK852498 KDR22680.1 AAZX01002341 AAZX01009423 AAZX01019112 GFDL01004078 JAV30967.1 GFDL01004070 JAV30975.1 GFDL01004071 JAV30974.1 GFDL01002874 JAV32171.1 GFDL01004052 JAV30993.1 GANO01003184 JAB56687.1 KQ971338 EFA02564.1 GFDL01004056 JAV30989.1 GFDL01002683 JAV32362.1 GFDL01002788 JAV32257.1 JR037114 AEY57590.1 GFDL01000910 JAV34135.1 GFDL01003732 JAV31313.1 GFDL01003710 JAV31335.1 GFDL01000771 JAV34274.1 GFDL01000833 JAV34212.1 GFDL01002803 JAV32242.1 GFDL01003718 JAV31327.1 GFDL01003714 JAV31331.1 GFDL01002628 JAV32417.1 GFDL01000656 JAV34389.1 AAAB01008986 EAL38873.2 CH478283 EAT33324.1 GEZM01030030 JAV85833.1 JR037113 AEY57589.1 GEZM01030028 JAV85834.1 GAPW01001217 JAC12381.1 GECZ01004170 JAS65599.1 GECU01017164 JAS90542.1 KK854114 PTY11524.1 GEZM01030035 JAV85828.1 OUUW01000001 SPP73755.1 GEZM01030033 JAV85830.1 KA646698 AFP61327.1 CH940659 KRF85706.1 CM000071 KRT02346.1 CH933808 EDW10857.2 CH902619 EDV37910.2 EZ422829 ADD19105.1 CH954179 EDV56156.2 KQS62729.1 GGFK01004714 MBW38035.1 DS235833 EEB18154.1 GDKW01002240 JAI54355.1 GBXI01007290 JAD07002.1 GEBQ01023761 GEBQ01021301 GEBQ01019585 JAT16216.1 JAT18676.1 JAT20392.1 GEBQ01011538 JAT28439.1 ACPB03023223 GAHY01001559 JAA75951.1 CM000158 EDW90609.2 GDHF01031185 GDHF01015416 GDHF01011730 JAI21129.1 JAI36898.1 JAI40584.1 GAKP01010888 GAKP01010886 JAC48066.1 CM002911 KMY93515.1 GECZ01012534 JAS57235.1 GDAI01000230 JAI17373.1 SPP73756.1 EAL25531.3 AE013599 AAF58369.1 BT150228 AGU12789.1 EDW71406.2 GBHO01007049 JAG36555.1 KPU77100.1 GBGD01001661 JAC87228.1 GBHO01039100 GBRD01000766 JAG04504.1 JAG65055.1 GECZ01007069 JAS62700.1 SPP73754.1 KRT02345.1 GECL01003409 JAP02715.1 GECZ01031118 JAS38651.1 KQS62731.1 GDHF01010456 GDHF01000743 JAI41858.1 JAI51571.1 GAKP01010887 GAKP01010883 JAC48069.1 GAMC01018758 JAB87797.1 GBXI01001304 GBXI01000381 JAD12988.1 JAD13911.1 KRJ99320.1 GFDF01002653 JAV11431.1 KMY93513.1 GEBQ01023569 JAT16408.1 GAKP01010885 GAKP01010884 JAC48068.1 AAF58370.1 KPU77101.1 GDHF01012387 GDHF01004636 GDHF01002563 JAI39927.1 JAI47678.1 JAI49751.1
Proteomes
Pfam
PF01066 CDP-OH_P_transf
ProteinModelPortal
H9IUZ3
A0A2A4J7T5
A0A2A4J6I1
A0A2A4J7J8
A0A3S2LSD2
A0A026WR91
+ More
A0A2J7PMM8 A0A067RQ11 K7J4I6 A0A1Q3FTS1 A0A1Q3FTT4 A0A1Q3FTQ2 A0A1Q3FX27 A0A1Q3FU90 U5ESN6 D2A0U9 A0A1Q3FTQ8 A0A1Q3FY07 A0A1Q3FXE0 V9I944 A0A1Q3G2T6 A0A1Q3FV56 A0A1Q3FUW0 A0A1Q3G397 A0A1Q3G329 A0A1Q3FXP1 A0A1Q3FUT5 A0A1Q3FUQ2 A0A1Q3FXX3 A0A1Q3G3Q2 Q5TMM4 Q16GF6 A0A1Y1MJH0 V9IBC0 A0A1Y1MJD2 A0A023EU02 A0A1W4X2U1 A0A1B6GT41 A0A1B6IUG1 A0A2R7VVK9 A0A1Y1ML92 A0A3B0IZQ9 A0A1W4X1Q6 A0A1Y1MMA7 T1PFL7 A0A0Q9WLY1 A0A1I8PM53 A0A0R3NN33 B4KML0 A0A1W4WS27 B3MGZ2 A0A1W4V4E3 D3TN27 B3NRK4 A0A2M4AB71 E0VXP8 A0A0P4VN76 A0A0A1X798 A0A1B6L9K1 A0A1B6LXK0 R4FMY2 B4P4H0 A0A0K8VNW2 A0A034VXL9 A0A0J9U111 A0A1B6G466 A0A0K8TT93 A0A3B0JKI2 Q28ZT2 A1Z9E0 U3GRY8 A0A1I8PM90 A0A1I8N2M4 B4MCL8 A0A0A9Z421 A0A0N8P0H0 A0A069DT05 A0A0A9W9M4 A0A1B6GJY7 A0A3B0IZG4 A0A0R3NN50 A0A0V0G3U8 A0A1B6EL77 A0A1I8N2N6 A0A1W4VGM2 A0A0Q5VX37 A0A0K8WK95 A0A1I8PM80 A0A034W061 W8AP52 A0A0A1XQY5 A0A0R1DQ53 A0A1L8DYA7 A0A0J9RBY5 A0A1B6KY80 A0A034W2R5 A0A1I8N2R1 A1Z9D9 A0A0P8ZRN5 A0A0K8W8Z9
A0A2J7PMM8 A0A067RQ11 K7J4I6 A0A1Q3FTS1 A0A1Q3FTT4 A0A1Q3FTQ2 A0A1Q3FX27 A0A1Q3FU90 U5ESN6 D2A0U9 A0A1Q3FTQ8 A0A1Q3FY07 A0A1Q3FXE0 V9I944 A0A1Q3G2T6 A0A1Q3FV56 A0A1Q3FUW0 A0A1Q3G397 A0A1Q3G329 A0A1Q3FXP1 A0A1Q3FUT5 A0A1Q3FUQ2 A0A1Q3FXX3 A0A1Q3G3Q2 Q5TMM4 Q16GF6 A0A1Y1MJH0 V9IBC0 A0A1Y1MJD2 A0A023EU02 A0A1W4X2U1 A0A1B6GT41 A0A1B6IUG1 A0A2R7VVK9 A0A1Y1ML92 A0A3B0IZQ9 A0A1W4X1Q6 A0A1Y1MMA7 T1PFL7 A0A0Q9WLY1 A0A1I8PM53 A0A0R3NN33 B4KML0 A0A1W4WS27 B3MGZ2 A0A1W4V4E3 D3TN27 B3NRK4 A0A2M4AB71 E0VXP8 A0A0P4VN76 A0A0A1X798 A0A1B6L9K1 A0A1B6LXK0 R4FMY2 B4P4H0 A0A0K8VNW2 A0A034VXL9 A0A0J9U111 A0A1B6G466 A0A0K8TT93 A0A3B0JKI2 Q28ZT2 A1Z9E0 U3GRY8 A0A1I8PM90 A0A1I8N2M4 B4MCL8 A0A0A9Z421 A0A0N8P0H0 A0A069DT05 A0A0A9W9M4 A0A1B6GJY7 A0A3B0IZG4 A0A0R3NN50 A0A0V0G3U8 A0A1B6EL77 A0A1I8N2N6 A0A1W4VGM2 A0A0Q5VX37 A0A0K8WK95 A0A1I8PM80 A0A034W061 W8AP52 A0A0A1XQY5 A0A0R1DQ53 A0A1L8DYA7 A0A0J9RBY5 A0A1B6KY80 A0A034W2R5 A0A1I8N2R1 A1Z9D9 A0A0P8ZRN5 A0A0K8W8Z9
Ontologies
GO
PANTHER
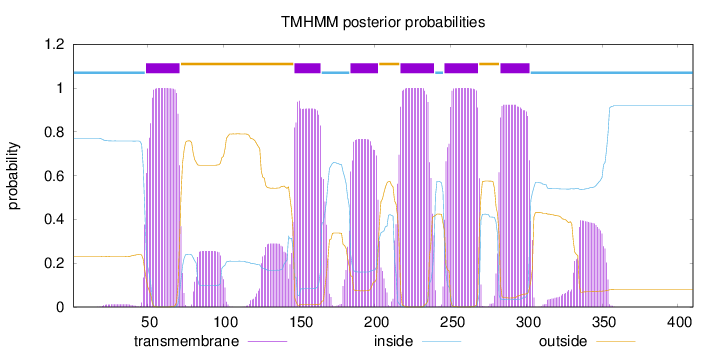
Topology
Length:
410
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
141.41017
Exp number, first 60 AAs:
11.65952
Total prob of N-in:
0.76966
POSSIBLE N-term signal
sequence
inside
1 - 48
TMhelix
49 - 71
outside
72 - 146
TMhelix
147 - 164
inside
165 - 183
TMhelix
184 - 202
outside
203 - 216
TMhelix
217 - 239
inside
240 - 245
TMhelix
246 - 268
outside
269 - 282
TMhelix
283 - 302
inside
303 - 410
Population Genetic Test Statistics
Pi
23.517547
Theta
21.15534
Tajima's D
0.112757
CLR
1.1627
CSRT
0.404979751012449
Interpretation
Uncertain
