Pre Gene Modal
BGIBMGA001070
Annotation
ADP-ribosylation_factor-like_protein_1_[Spodoptera_exigua]
Full name
ADP-ribosylation factor-like protein 1
Location in the cell
PlasmaMembrane Reliability : 0.937
Sequence
CDS
ATGGGTGGTCTATTTAGTTACTTTAGAGGATTGCTGGGTGCAAGGGAAATGCGGATTTTGATACTTGGATTGGACGGTGCCGGAAAGACGACAATTCTTTATAAATTGCAAGTCGGTGAGGTCGTGACCACTATACCGACGATTGGGTTCAACGTAGAACAAGTCACATATAAAAATCTCAAGTTCCAAGTGTGGGATCTTGGGGGACAGACCAGTATTAGGCCTTATTGGCGATGTTACTATGGCAACACAGATGCAATAATATATGTTGTAGACTCAGCTGACAGGGATCGAATAGGAATATCTAAAGATGAACTTGTGCATATGTTAAGGGAAGAAGAATTAGCGAACGCGATACTCGTTGTTTTAGCCAACAAACAGGACATGGCCGGATGTCTGACAGTAGCCGAGGTACACCAGGCCCTGGGCTTGGACGCCTTGCGCGATAGAACCTTCCAGATCTTTAAGACTTCAGCCGTCAGAGGCGAGGGGCTAGATCAGGCGATGGACTGGCTCTCGAACGCGCTTCAAGCTAGGAAATAA
Protein
MGGLFSYFRGLLGAREMRILILGLDGAGKTTILYKLQVGEVVTTIPTIGFNVEQVTYKNLKFQVWDLGGQTSIRPYWRCYYGNTDAIIYVVDSADRDRIGISKDELVHMLREEELANAILVVLANKQDMAGCLTVAEVHQALGLDALRDRTFQIFKTSAVRGEGLDQAMDWLSNALQARK
Summary
Description
GTP-binding protein involved in protein trafficking; may modulate vesicle budding and uncoating within the Golgi apparatus.
Similarity
Belongs to the small GTPase superfamily. Arf family.
Keywords
Complete proteome
GTP-binding
Lipoprotein
Myristate
Nucleotide-binding
Reference proteome
Feature
chain ADP-ribosylation factor-like protein 1
Uniprot
A0A2W1C262
A0A2A4JAB6
A0A3S2NB18
A0A1E1WDX3
I6R5P2
I4DNP4
+ More
S4PVG6 A0A0K8TN83 A0A182YL45 A0A1S4H8H4 A0A2M3ZJG6 A0A2M4AKT9 A0A182P6Y0 A0A182MWX4 A0A182Q5P8 A0A182S5S6 A0A182UZ06 W5JEU0 A0A182JZQ0 A0A182TPL5 A0A182WEC9 A0A182X6N6 A0A182L4K2 A0A182IJX6 A0A182FKP3 A0A182R4F0 A0A182HH41 A0A2A3EL73 A0A087ZVS0 A0A182NPP9 T1DTU9 Q7PZ49 A0A1W4XW95 F4WAJ9 T1E331 Q16H28 A0A023EID3 A0A026WH95 K7J4F1 A0A1Q3F8T2 E2B053 A0A1Q3F8I0 A0A154NW44 A0A336KFT1 A0A195E6S1 A0A151XGC7 B4KYI0 A0A1B0DLY1 A0A1Y1KXC3 U5EV55 A0A1Q3F8R4 A0A1J1HPC9 W8C216 A0A3B0J583 A0A0K8W6K0 Q29D91 B4H7T6 A0A034W5R3 B4N724 A0A0A1WNP2 A0A1B6C8Q6 B0W2T2 A0A1W4VP58 B4ITV7 B3M9U1 B4HIL9 B4J3P0 B3NDP9 B4QLS2 P25160 B4LDZ5 A0A1I8M3I7 A0A1I8NV92 A0A0L0CRW2 A0A1L8EH66 A0A0T6AU96 A0A1L8DVV0 A0A0A9WX31 H9IUZ0 A0A212EVL6 A0A165VGC6 A0A195F8N6 A0A151I3L6 A0A1B0FBR7 A0A1A9XT25 A0A1A9VAR0 A0A1B0B965 A0A1B0AIE8 A0A194RG05 A0A1A9WGH4 A0A0M4EBG0 A0A1B6F4J4 A0A1B6JST1 A0A2P8Z5V2 A0A1B6MI19 A0A067RIZ8 A0A2J7PQ98 A0A2R7VWF0 A0A0P6AG63 E9G5T2
S4PVG6 A0A0K8TN83 A0A182YL45 A0A1S4H8H4 A0A2M3ZJG6 A0A2M4AKT9 A0A182P6Y0 A0A182MWX4 A0A182Q5P8 A0A182S5S6 A0A182UZ06 W5JEU0 A0A182JZQ0 A0A182TPL5 A0A182WEC9 A0A182X6N6 A0A182L4K2 A0A182IJX6 A0A182FKP3 A0A182R4F0 A0A182HH41 A0A2A3EL73 A0A087ZVS0 A0A182NPP9 T1DTU9 Q7PZ49 A0A1W4XW95 F4WAJ9 T1E331 Q16H28 A0A023EID3 A0A026WH95 K7J4F1 A0A1Q3F8T2 E2B053 A0A1Q3F8I0 A0A154NW44 A0A336KFT1 A0A195E6S1 A0A151XGC7 B4KYI0 A0A1B0DLY1 A0A1Y1KXC3 U5EV55 A0A1Q3F8R4 A0A1J1HPC9 W8C216 A0A3B0J583 A0A0K8W6K0 Q29D91 B4H7T6 A0A034W5R3 B4N724 A0A0A1WNP2 A0A1B6C8Q6 B0W2T2 A0A1W4VP58 B4ITV7 B3M9U1 B4HIL9 B4J3P0 B3NDP9 B4QLS2 P25160 B4LDZ5 A0A1I8M3I7 A0A1I8NV92 A0A0L0CRW2 A0A1L8EH66 A0A0T6AU96 A0A1L8DVV0 A0A0A9WX31 H9IUZ0 A0A212EVL6 A0A165VGC6 A0A195F8N6 A0A151I3L6 A0A1B0FBR7 A0A1A9XT25 A0A1A9VAR0 A0A1B0B965 A0A1B0AIE8 A0A194RG05 A0A1A9WGH4 A0A0M4EBG0 A0A1B6F4J4 A0A1B6JST1 A0A2P8Z5V2 A0A1B6MI19 A0A067RIZ8 A0A2J7PQ98 A0A2R7VWF0 A0A0P6AG63 E9G5T2
Pubmed
28756777
22651552
23622113
26369729
25244985
12364791
+ More
20920257 23761445 20966253 21719571 24330624 17510324 24945155 26483478 24508170 30249741 20075255 20798317 17994087 28004739 24495485 15632085 25348373 25830018 17550304 22936249 1901655 10731132 12537572 12537569 25315136 26108605 25401762 26823975 19121390 22118469 26354079 29403074 24845553 21292972
20920257 23761445 20966253 21719571 24330624 17510324 24945155 26483478 24508170 30249741 20075255 20798317 17994087 28004739 24495485 15632085 25348373 25830018 17550304 22936249 1901655 10731132 12537572 12537569 25315136 26108605 25401762 26823975 19121390 22118469 26354079 29403074 24845553 21292972
EMBL
KZ149891
PZC79066.1
NWSH01002411
PCG68343.1
RSAL01000294
RVE42853.1
+ More
GDQN01005993 JAT85061.1 JQ653045 AFM38217.1 AK403063 BAM19534.1 GAIX01008573 JAA83987.1 GDAI01001779 JAI15824.1 AAAB01008986 GGFM01007874 MBW28625.1 GGFK01008053 MBW41374.1 AXCM01000779 AXCN02001661 ADMH02001696 ETN61395.1 AXCP01005566 APCN01002384 KZ288219 PBC32244.1 GAMD01000739 JAB00852.1 EAA00052.3 GL888050 EGI68757.1 GALA01000740 JAA94112.1 CH478211 EAT33549.1 JXUM01116539 JXUM01116540 GAPW01004908 KQ565995 JAC08690.1 KXJ70468.1 KK107211 QOIP01000005 EZA55333.1 RLU22695.1 GFDL01011083 JAV23962.1 GL444392 EFN60938.1 GFDL01011171 JAV23874.1 KQ434772 KZC03867.1 UFQS01000115 UFQT01000115 SSW99920.1 SSX20300.1 KQ979592 KYN20544.1 KQ982174 KYQ59300.1 CH933809 EDW18791.1 AJVK01036729 GEZM01076957 JAV63517.1 GANO01001173 JAB58698.1 GFDL01011109 JAV23936.1 CVRI01000015 CRK89903.1 GAMC01000688 JAC05868.1 OUUW01000002 SPP76795.1 GDHF01005630 JAI46684.1 CH379070 EAL30523.1 CH479219 EDW34726.1 GAKP01008031 JAC50921.1 CH964168 EDW80163.2 GBXI01013840 JAD00452.1 GEDC01027663 JAS09635.1 DS231828 EDS29835.1 CH891734 EDW99820.1 CH902618 EDV40132.1 CH480815 EDW41649.1 CH916366 EDV97271.1 CH954178 EDV52182.2 CM000363 CM002912 EDX10621.1 KMY99890.1 M61127 AE014296 BT001460 CH940647 EDW70038.1 JRES01000005 KNC34961.1 GFDG01000768 JAV18031.1 LJIG01022780 KRT78752.1 GFDF01003532 JAV10552.1 GBHO01030597 GBRD01005309 GDHC01021093 JAG13007.1 JAG60512.1 JAP97535.1 BABH01000062 BABH01000063 AGBW02012175 OWR45532.1 KT984815 AMZ00367.1 KQ981727 KYN36741.1 KQ976484 KYM83689.1 CCAG010017145 JXJN01010286 KQ460297 KPJ16240.1 CP012525 ALC44791.1 GECZ01024661 JAS45108.1 GECU01005489 JAT02218.1 PYGN01000183 PSN51867.1 GEBQ01004436 JAT35541.1 KK852442 KDR23846.1 NEVH01022640 PNF18518.1 KK854132 PTY11826.1 GDIP01043163 JAM60552.1 GL732533 EFX85116.1
GDQN01005993 JAT85061.1 JQ653045 AFM38217.1 AK403063 BAM19534.1 GAIX01008573 JAA83987.1 GDAI01001779 JAI15824.1 AAAB01008986 GGFM01007874 MBW28625.1 GGFK01008053 MBW41374.1 AXCM01000779 AXCN02001661 ADMH02001696 ETN61395.1 AXCP01005566 APCN01002384 KZ288219 PBC32244.1 GAMD01000739 JAB00852.1 EAA00052.3 GL888050 EGI68757.1 GALA01000740 JAA94112.1 CH478211 EAT33549.1 JXUM01116539 JXUM01116540 GAPW01004908 KQ565995 JAC08690.1 KXJ70468.1 KK107211 QOIP01000005 EZA55333.1 RLU22695.1 GFDL01011083 JAV23962.1 GL444392 EFN60938.1 GFDL01011171 JAV23874.1 KQ434772 KZC03867.1 UFQS01000115 UFQT01000115 SSW99920.1 SSX20300.1 KQ979592 KYN20544.1 KQ982174 KYQ59300.1 CH933809 EDW18791.1 AJVK01036729 GEZM01076957 JAV63517.1 GANO01001173 JAB58698.1 GFDL01011109 JAV23936.1 CVRI01000015 CRK89903.1 GAMC01000688 JAC05868.1 OUUW01000002 SPP76795.1 GDHF01005630 JAI46684.1 CH379070 EAL30523.1 CH479219 EDW34726.1 GAKP01008031 JAC50921.1 CH964168 EDW80163.2 GBXI01013840 JAD00452.1 GEDC01027663 JAS09635.1 DS231828 EDS29835.1 CH891734 EDW99820.1 CH902618 EDV40132.1 CH480815 EDW41649.1 CH916366 EDV97271.1 CH954178 EDV52182.2 CM000363 CM002912 EDX10621.1 KMY99890.1 M61127 AE014296 BT001460 CH940647 EDW70038.1 JRES01000005 KNC34961.1 GFDG01000768 JAV18031.1 LJIG01022780 KRT78752.1 GFDF01003532 JAV10552.1 GBHO01030597 GBRD01005309 GDHC01021093 JAG13007.1 JAG60512.1 JAP97535.1 BABH01000062 BABH01000063 AGBW02012175 OWR45532.1 KT984815 AMZ00367.1 KQ981727 KYN36741.1 KQ976484 KYM83689.1 CCAG010017145 JXJN01010286 KQ460297 KPJ16240.1 CP012525 ALC44791.1 GECZ01024661 JAS45108.1 GECU01005489 JAT02218.1 PYGN01000183 PSN51867.1 GEBQ01004436 JAT35541.1 KK852442 KDR23846.1 NEVH01022640 PNF18518.1 KK854132 PTY11826.1 GDIP01043163 JAM60552.1 GL732533 EFX85116.1
Proteomes
UP000218220
UP000283053
UP000076408
UP000075885
UP000075883
UP000075886
+ More
UP000075901 UP000075903 UP000000673 UP000075881 UP000075902 UP000075920 UP000076407 UP000075882 UP000075880 UP000069272 UP000075900 UP000075840 UP000242457 UP000005203 UP000075884 UP000007062 UP000192223 UP000007755 UP000008820 UP000069940 UP000249989 UP000053097 UP000279307 UP000002358 UP000000311 UP000076502 UP000078492 UP000075809 UP000009192 UP000092462 UP000183832 UP000268350 UP000001819 UP000008744 UP000007798 UP000002320 UP000192221 UP000002282 UP000007801 UP000001292 UP000001070 UP000008711 UP000000304 UP000000803 UP000008792 UP000095301 UP000095300 UP000037069 UP000005204 UP000007151 UP000078541 UP000078540 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000053240 UP000091820 UP000092553 UP000245037 UP000027135 UP000235965 UP000000305
UP000075901 UP000075903 UP000000673 UP000075881 UP000075902 UP000075920 UP000076407 UP000075882 UP000075880 UP000069272 UP000075900 UP000075840 UP000242457 UP000005203 UP000075884 UP000007062 UP000192223 UP000007755 UP000008820 UP000069940 UP000249989 UP000053097 UP000279307 UP000002358 UP000000311 UP000076502 UP000078492 UP000075809 UP000009192 UP000092462 UP000183832 UP000268350 UP000001819 UP000008744 UP000007798 UP000002320 UP000192221 UP000002282 UP000007801 UP000001292 UP000001070 UP000008711 UP000000304 UP000000803 UP000008792 UP000095301 UP000095300 UP000037069 UP000005204 UP000007151 UP000078541 UP000078540 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000053240 UP000091820 UP000092553 UP000245037 UP000027135 UP000235965 UP000000305
Pfam
PF00025 Arf
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2W1C262
A0A2A4JAB6
A0A3S2NB18
A0A1E1WDX3
I6R5P2
I4DNP4
+ More
S4PVG6 A0A0K8TN83 A0A182YL45 A0A1S4H8H4 A0A2M3ZJG6 A0A2M4AKT9 A0A182P6Y0 A0A182MWX4 A0A182Q5P8 A0A182S5S6 A0A182UZ06 W5JEU0 A0A182JZQ0 A0A182TPL5 A0A182WEC9 A0A182X6N6 A0A182L4K2 A0A182IJX6 A0A182FKP3 A0A182R4F0 A0A182HH41 A0A2A3EL73 A0A087ZVS0 A0A182NPP9 T1DTU9 Q7PZ49 A0A1W4XW95 F4WAJ9 T1E331 Q16H28 A0A023EID3 A0A026WH95 K7J4F1 A0A1Q3F8T2 E2B053 A0A1Q3F8I0 A0A154NW44 A0A336KFT1 A0A195E6S1 A0A151XGC7 B4KYI0 A0A1B0DLY1 A0A1Y1KXC3 U5EV55 A0A1Q3F8R4 A0A1J1HPC9 W8C216 A0A3B0J583 A0A0K8W6K0 Q29D91 B4H7T6 A0A034W5R3 B4N724 A0A0A1WNP2 A0A1B6C8Q6 B0W2T2 A0A1W4VP58 B4ITV7 B3M9U1 B4HIL9 B4J3P0 B3NDP9 B4QLS2 P25160 B4LDZ5 A0A1I8M3I7 A0A1I8NV92 A0A0L0CRW2 A0A1L8EH66 A0A0T6AU96 A0A1L8DVV0 A0A0A9WX31 H9IUZ0 A0A212EVL6 A0A165VGC6 A0A195F8N6 A0A151I3L6 A0A1B0FBR7 A0A1A9XT25 A0A1A9VAR0 A0A1B0B965 A0A1B0AIE8 A0A194RG05 A0A1A9WGH4 A0A0M4EBG0 A0A1B6F4J4 A0A1B6JST1 A0A2P8Z5V2 A0A1B6MI19 A0A067RIZ8 A0A2J7PQ98 A0A2R7VWF0 A0A0P6AG63 E9G5T2
S4PVG6 A0A0K8TN83 A0A182YL45 A0A1S4H8H4 A0A2M3ZJG6 A0A2M4AKT9 A0A182P6Y0 A0A182MWX4 A0A182Q5P8 A0A182S5S6 A0A182UZ06 W5JEU0 A0A182JZQ0 A0A182TPL5 A0A182WEC9 A0A182X6N6 A0A182L4K2 A0A182IJX6 A0A182FKP3 A0A182R4F0 A0A182HH41 A0A2A3EL73 A0A087ZVS0 A0A182NPP9 T1DTU9 Q7PZ49 A0A1W4XW95 F4WAJ9 T1E331 Q16H28 A0A023EID3 A0A026WH95 K7J4F1 A0A1Q3F8T2 E2B053 A0A1Q3F8I0 A0A154NW44 A0A336KFT1 A0A195E6S1 A0A151XGC7 B4KYI0 A0A1B0DLY1 A0A1Y1KXC3 U5EV55 A0A1Q3F8R4 A0A1J1HPC9 W8C216 A0A3B0J583 A0A0K8W6K0 Q29D91 B4H7T6 A0A034W5R3 B4N724 A0A0A1WNP2 A0A1B6C8Q6 B0W2T2 A0A1W4VP58 B4ITV7 B3M9U1 B4HIL9 B4J3P0 B3NDP9 B4QLS2 P25160 B4LDZ5 A0A1I8M3I7 A0A1I8NV92 A0A0L0CRW2 A0A1L8EH66 A0A0T6AU96 A0A1L8DVV0 A0A0A9WX31 H9IUZ0 A0A212EVL6 A0A165VGC6 A0A195F8N6 A0A151I3L6 A0A1B0FBR7 A0A1A9XT25 A0A1A9VAR0 A0A1B0B965 A0A1B0AIE8 A0A194RG05 A0A1A9WGH4 A0A0M4EBG0 A0A1B6F4J4 A0A1B6JST1 A0A2P8Z5V2 A0A1B6MI19 A0A067RIZ8 A0A2J7PQ98 A0A2R7VWF0 A0A0P6AG63 E9G5T2
PDB
1R4A
E-value=8.39637e-72,
Score=682
Ontologies
GO
GO:0005525
GO:0016192
GO:0005737
GO:0005794
GO:0006886
GO:0090158
GO:0030334
GO:0060628
GO:0019904
GO:0007030
GO:0007431
GO:0035220
GO:1903292
GO:0070861
GO:0033363
GO:0005802
GO:0033227
GO:0048488
GO:0007269
GO:0003924
GO:0098793
GO:0034067
GO:0005622
GO:0007264
GO:0016992
GO:0051539
GO:0015914
GO:0046872
GO:0006412
GO:0016876
GO:0043039
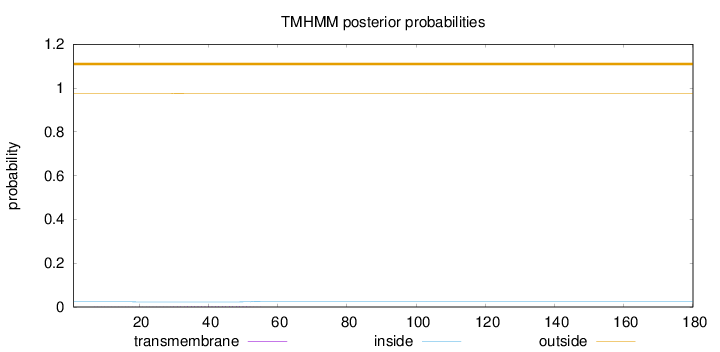
Topology
Length:
180
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06265
Exp number, first 60 AAs:
0.06199
Total prob of N-in:
0.02352
outside
1 - 180
Population Genetic Test Statistics
Pi
21.843193
Theta
17.626519
Tajima's D
-0.829839
CLR
13.933055
CSRT
0.166641667916604
Interpretation
Uncertain
