Gene
KWMTBOMO08129 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001092
Annotation
PREDICTED:_uncharacterized_protein_LOC101735793_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.285
Sequence
CDS
ATGGAGAAGCTCGTTCTGTGTTTAGTGTTGTTTGCGGTTGTAGCCGTGCAAGCGAGACACCAGTTCCCTAGAGTCAGCGAGCGCCTTGGGGAACCCTTCAGGATAGGGGAATACATCAAAGTTGTCTGCACGTCAGAACTCAGCAACGAAAGGGATCACATCATGGAAAAGTCGAGATGCGGTGAACCGAAGGATGTCTTCGTGCAGTTAAAACCTAACTCACCTCACGAGCTGGTGAGCCCTAGCGCGGTTTGGGTGAAGAACTGTGTCGGGATTTGCGATTATGACAATGAAGGATCCTGTATTGCGACTGAAACAAAAATCAGACACATACCGGTACGAATATACAATGGAAATACGAATAAAGAATCATGTGCCACGTACGCGGTGGAGGAGCACGTCAAGTGTGGGTGCTGCGCCCTGAGTCCCGACCACTGCGTCTCCCCCAGGGTGTTCAATCCTCGTAAGTGCAGCTGCCACTGCCCCAATATCGAGGACAGGAGGGAATGCGTCAAGAAGACGGACATGACTTGGAACCGAGCTCACTGCAAATGTGAAAAGATATAG
Protein
MEKLVLCLVLFAVVAVQARHQFPRVSERLGEPFRIGEYIKVVCTSELSNERDHIMEKSRCGEPKDVFVQLKPNSPHELVSPSAVWVKNCVGICDYDNEGSCIATETKIRHIPVRIYNGNTNKESCATYAVEEHVKCGCCALSPDHCVSPRVFNPRKCSCHCPNIEDRRECVKKTDMTWNRAHCKCEKI
Summary
Similarity
Belongs to the PDGF/VEGF growth factor family.
Uniprot
H9IV12
A0A0L7KWS4
A0A2W1C0P6
A0A2H1W2J3
A0A194RFK4
A0A194Q691
+ More
A0A212EVM2 A0A3S2P6I2 A0A2J7Q1V3 A0A067QJT5 A0A2P8YED6 A0A195D3H7 A0A158P1N2 A0A195DC45 A0A2P8Z536 A0A226E814 A0A182SXT9 A0A1Y1NK43 A0A1Y1NGK6 A0A182Y7H6 A0A3L8D7M3 A0A0K2UYJ7 A0A2S2QPU2 A0A026W4B9 C4WS19 J9PD10 A0A2M4CY31 A0A2M3Z865 A0A084VAA4 A0A2M3ZID6 A0A2M3Z8Q8 A0A2M3Z7I6 A0A2M4BTE5 A0A2M4BT88 C4WS18 A0A2M3ZI38 A0A2M3ZCE0 A0A182W4R0 A0A2M3ZCD1 A0A1W4XDN0 A0A2M3ZE55 W5JKH3 A0A2M4CZG2 A0A182RV46 A0A182M748 A0A2M4AM54 A0A2M4CZC9 A0A2M4CLX2 A0A182FIP6 A0A2M4AIP1 A0A2M4CHK2 A0A2M4CHP4 A0A2M4AMA0 A0A2D1N520 A0A3G2KWW0 A0A2M4AK81 B0X067 A0A2M4ARU3 A0A2M4AM82 A0A1D2N837 A0A034VBN1 A0A182J870 A0A034VD83 A0A182QAG6 A0A182IA60 A0A154NZG5 A0A182NVH4 A0A182L7Z4
A0A212EVM2 A0A3S2P6I2 A0A2J7Q1V3 A0A067QJT5 A0A2P8YED6 A0A195D3H7 A0A158P1N2 A0A195DC45 A0A2P8Z536 A0A226E814 A0A182SXT9 A0A1Y1NK43 A0A1Y1NGK6 A0A182Y7H6 A0A3L8D7M3 A0A0K2UYJ7 A0A2S2QPU2 A0A026W4B9 C4WS19 J9PD10 A0A2M4CY31 A0A2M3Z865 A0A084VAA4 A0A2M3ZID6 A0A2M3Z8Q8 A0A2M3Z7I6 A0A2M4BTE5 A0A2M4BT88 C4WS18 A0A2M3ZI38 A0A2M3ZCE0 A0A182W4R0 A0A2M3ZCD1 A0A1W4XDN0 A0A2M3ZE55 W5JKH3 A0A2M4CZG2 A0A182RV46 A0A182M748 A0A2M4AM54 A0A2M4CZC9 A0A2M4CLX2 A0A182FIP6 A0A2M4AIP1 A0A2M4CHK2 A0A2M4CHP4 A0A2M4AMA0 A0A2D1N520 A0A3G2KWW0 A0A2M4AK81 B0X067 A0A2M4ARU3 A0A2M4AM82 A0A1D2N837 A0A034VBN1 A0A182J870 A0A034VD83 A0A182QAG6 A0A182IA60 A0A154NZG5 A0A182NVH4 A0A182L7Z4
Pubmed
EMBL
BABH01000064
BABH01000065
BABH01000066
BABH01000067
JTDY01004935
KOB67580.1
+ More
KZ149891 PZC79067.1 ODYU01005883 SOQ47233.1 KQ460297 KPJ16239.1 KQ459439 KPJ01053.1 AGBW02012175 OWR45533.1 RSAL01000286 RVE42962.1 NEVH01019377 PNF22566.1 KK853392 KDR07915.1 PYGN01000661 PSN42629.1 KQ976948 KYN06924.1 ADTU01001092 ADTU01001093 KQ980989 KYN10485.1 PYGN01000191 PSN51614.1 LNIX01000006 OXA53127.1 GEZM01003020 JAV96975.1 GEZM01003024 JAV96971.1 QOIP01000012 RLU16299.1 HACA01025556 CDW42917.1 GGMS01010490 MBY79693.1 KK107488 EZA49889.1 AK340012 BAH70689.1 GU002536 ADF87936.1 GGFL01006064 MBW70242.1 GGFM01003929 MBW24680.1 ATLV01003052 KE524109 KFB34898.1 GGFM01007532 MBW28283.1 GGFM01004165 MBW24916.1 GGFM01003723 MBW24474.1 GGFJ01007133 MBW56274.1 GGFJ01007139 MBW56280.1 AK340011 BAH70688.1 GGFM01007384 MBW28135.1 GGFM01005412 MBW26163.1 GGFM01005384 MBW26135.1 GGFM01006010 MBW26761.1 ADMH02000934 ETN64636.1 GGFL01006532 MBW70710.1 AXCM01001248 GGFK01008544 MBW41865.1 GGFL01006508 MBW70686.1 GGFL01002101 MBW66279.1 GGFK01007338 MBW40659.1 GGFL01000605 MBW64783.1 GGFL01000577 MBW64755.1 GGFK01008605 MBW41926.1 KY356888 ATO74511.1 MH475138 QCYY01000154 AYN64309.1 ROT85909.1 GGFK01007869 MBW41190.1 DS232229 EDS37942.1 GGFK01010168 MBW43489.1 GGFK01008585 MBW41906.1 LJIJ01000169 ODN01136.1 GAKP01019430 JAC39522.1 AXCP01007866 AXCP01007867 AXCP01007868 GAKP01019429 JAC39523.1 AXCN02002155 APCN01001124 KQ434778 KZC04388.1
KZ149891 PZC79067.1 ODYU01005883 SOQ47233.1 KQ460297 KPJ16239.1 KQ459439 KPJ01053.1 AGBW02012175 OWR45533.1 RSAL01000286 RVE42962.1 NEVH01019377 PNF22566.1 KK853392 KDR07915.1 PYGN01000661 PSN42629.1 KQ976948 KYN06924.1 ADTU01001092 ADTU01001093 KQ980989 KYN10485.1 PYGN01000191 PSN51614.1 LNIX01000006 OXA53127.1 GEZM01003020 JAV96975.1 GEZM01003024 JAV96971.1 QOIP01000012 RLU16299.1 HACA01025556 CDW42917.1 GGMS01010490 MBY79693.1 KK107488 EZA49889.1 AK340012 BAH70689.1 GU002536 ADF87936.1 GGFL01006064 MBW70242.1 GGFM01003929 MBW24680.1 ATLV01003052 KE524109 KFB34898.1 GGFM01007532 MBW28283.1 GGFM01004165 MBW24916.1 GGFM01003723 MBW24474.1 GGFJ01007133 MBW56274.1 GGFJ01007139 MBW56280.1 AK340011 BAH70688.1 GGFM01007384 MBW28135.1 GGFM01005412 MBW26163.1 GGFM01005384 MBW26135.1 GGFM01006010 MBW26761.1 ADMH02000934 ETN64636.1 GGFL01006532 MBW70710.1 AXCM01001248 GGFK01008544 MBW41865.1 GGFL01006508 MBW70686.1 GGFL01002101 MBW66279.1 GGFK01007338 MBW40659.1 GGFL01000605 MBW64783.1 GGFL01000577 MBW64755.1 GGFK01008605 MBW41926.1 KY356888 ATO74511.1 MH475138 QCYY01000154 AYN64309.1 ROT85909.1 GGFK01007869 MBW41190.1 DS232229 EDS37942.1 GGFK01010168 MBW43489.1 GGFK01008585 MBW41906.1 LJIJ01000169 ODN01136.1 GAKP01019430 JAC39522.1 AXCP01007866 AXCP01007867 AXCP01007868 GAKP01019429 JAC39523.1 AXCN02002155 APCN01001124 KQ434778 KZC04388.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000235965 UP000027135 UP000245037 UP000078542 UP000005205 UP000078492 UP000198287 UP000075901 UP000076408 UP000279307 UP000053097 UP000030765 UP000075920 UP000192223 UP000000673 UP000075900 UP000075883 UP000069272 UP000283509 UP000002320 UP000094527 UP000075880 UP000075886 UP000075840 UP000076502 UP000075884 UP000075882
UP000235965 UP000027135 UP000245037 UP000078542 UP000005205 UP000078492 UP000198287 UP000075901 UP000076408 UP000279307 UP000053097 UP000030765 UP000075920 UP000192223 UP000000673 UP000075900 UP000075883 UP000069272 UP000283509 UP000002320 UP000094527 UP000075880 UP000075886 UP000075840 UP000076502 UP000075884 UP000075882
PRIDE
Interpro
SUPFAM
SSF57501
SSF57501
Gene 3D
ProteinModelPortal
H9IV12
A0A0L7KWS4
A0A2W1C0P6
A0A2H1W2J3
A0A194RFK4
A0A194Q691
+ More
A0A212EVM2 A0A3S2P6I2 A0A2J7Q1V3 A0A067QJT5 A0A2P8YED6 A0A195D3H7 A0A158P1N2 A0A195DC45 A0A2P8Z536 A0A226E814 A0A182SXT9 A0A1Y1NK43 A0A1Y1NGK6 A0A182Y7H6 A0A3L8D7M3 A0A0K2UYJ7 A0A2S2QPU2 A0A026W4B9 C4WS19 J9PD10 A0A2M4CY31 A0A2M3Z865 A0A084VAA4 A0A2M3ZID6 A0A2M3Z8Q8 A0A2M3Z7I6 A0A2M4BTE5 A0A2M4BT88 C4WS18 A0A2M3ZI38 A0A2M3ZCE0 A0A182W4R0 A0A2M3ZCD1 A0A1W4XDN0 A0A2M3ZE55 W5JKH3 A0A2M4CZG2 A0A182RV46 A0A182M748 A0A2M4AM54 A0A2M4CZC9 A0A2M4CLX2 A0A182FIP6 A0A2M4AIP1 A0A2M4CHK2 A0A2M4CHP4 A0A2M4AMA0 A0A2D1N520 A0A3G2KWW0 A0A2M4AK81 B0X067 A0A2M4ARU3 A0A2M4AM82 A0A1D2N837 A0A034VBN1 A0A182J870 A0A034VD83 A0A182QAG6 A0A182IA60 A0A154NZG5 A0A182NVH4 A0A182L7Z4
A0A212EVM2 A0A3S2P6I2 A0A2J7Q1V3 A0A067QJT5 A0A2P8YED6 A0A195D3H7 A0A158P1N2 A0A195DC45 A0A2P8Z536 A0A226E814 A0A182SXT9 A0A1Y1NK43 A0A1Y1NGK6 A0A182Y7H6 A0A3L8D7M3 A0A0K2UYJ7 A0A2S2QPU2 A0A026W4B9 C4WS19 J9PD10 A0A2M4CY31 A0A2M3Z865 A0A084VAA4 A0A2M3ZID6 A0A2M3Z8Q8 A0A2M3Z7I6 A0A2M4BTE5 A0A2M4BT88 C4WS18 A0A2M3ZI38 A0A2M3ZCE0 A0A182W4R0 A0A2M3ZCD1 A0A1W4XDN0 A0A2M3ZE55 W5JKH3 A0A2M4CZG2 A0A182RV46 A0A182M748 A0A2M4AM54 A0A2M4CZC9 A0A2M4CLX2 A0A182FIP6 A0A2M4AIP1 A0A2M4CHK2 A0A2M4CHP4 A0A2M4AMA0 A0A2D1N520 A0A3G2KWW0 A0A2M4AK81 B0X067 A0A2M4ARU3 A0A2M4AM82 A0A1D2N837 A0A034VBN1 A0A182J870 A0A034VD83 A0A182QAG6 A0A182IA60 A0A154NZG5 A0A182NVH4 A0A182L7Z4
Ontologies
PANTHER
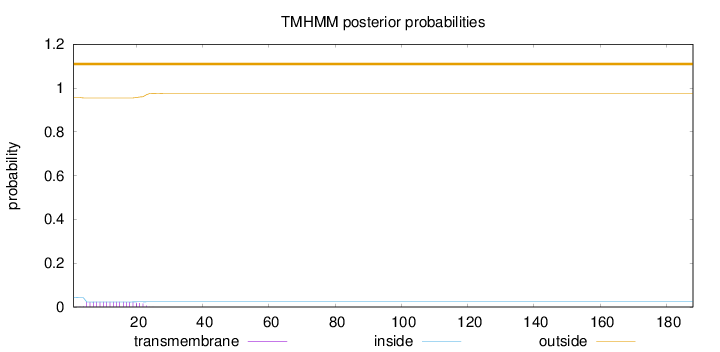
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.997754
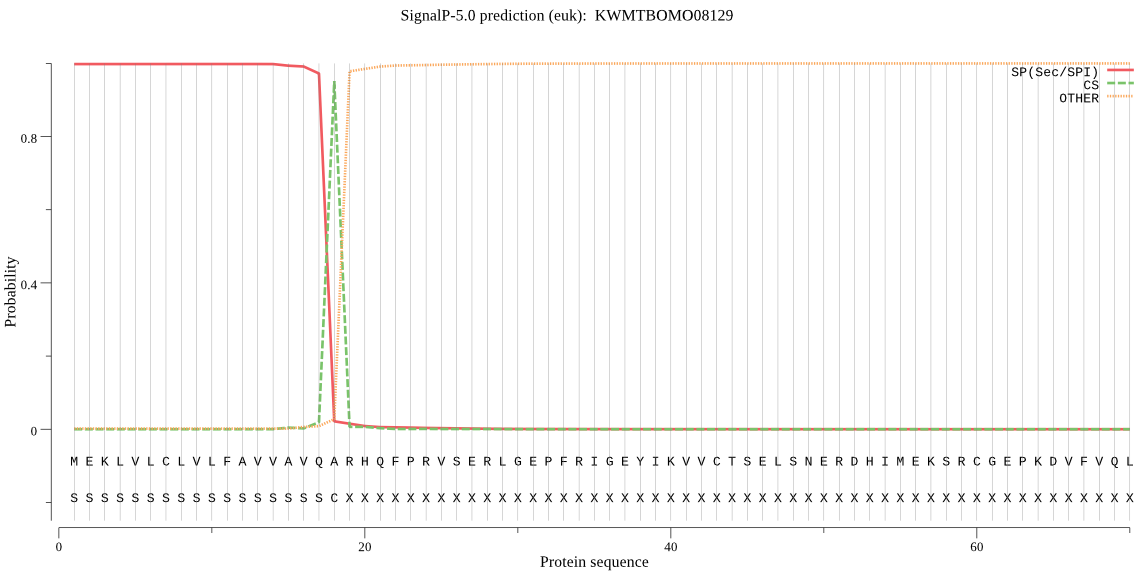
Length:
188
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.40915
Exp number, first 60 AAs:
0.40915
Total prob of N-in:
0.04383
outside
1 - 188
Population Genetic Test Statistics
Pi
19.665479
Theta
18.223447
Tajima's D
-0.926885
CLR
1.393544
CSRT
0.148792560371981
Interpretation
Uncertain
