Gene
KWMTBOMO08126 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001068
Annotation
Fumarylacetoacetate_hydrolase_isoform_A_[Operophtera_brumata]
Full name
Fumarylacetoacetate hydrolase domain-containing protein 2
Location in the cell
Cytoplasmic Reliability : 2.611
Sequence
CDS
ATGAAACTCCTACAGTTTTCTTACAAAAACAGCGATGGAATCCGAGCGGGTTTCTTAGATGGCGATAATGTTGTGGATATCAATAAAGAAGATCCAAAATTACCATCAACGTTAATTGAGCTCCTCAAAACGAATTCCATTTCCAAAGTGAAAAAAATCAGCGCCAAATCACCAGTGCCTTTATCCGGTATCACGCTGAAAGCTCCGGTCCATGGAAATGATAAAGTACTATGCGTGGGATTAAATTACAAAGATCACTGTGAAGAGCAGAAATTGACGCCTCCAGAACTTCCTTTTATATTCAACAAATTTCCTAGTACTGTGGTGGGACCAAACGACACCATCAAGTTAAAAATGGATGTAAGTAAAGCTGTGGTCTGCGAGATAGAACTGACGGTTGTAATCGGGAAGAAAGCCAGTAAAGTAGATTCATCGCATGCCTTTGATTATGTCCTCGGCTACACAATAGCCCAGGATATCGGCGCTACAGACTGGGAGAAGAACAAGAAAATCAGTCAATTGCTGTTGGGTAAAGCAATGGACACCTTCTGCCCAATTGGACCTTGGATAGTTACCAGCGATGAGATCGGAAATCCTCAGAATTTGAATGTAAAGTGCAGCATTAATGGCGTTCAGAAACAGAAGAGTAATACAAACCAATTTATCCACAAAATCCCGGATATTATTGCAAGATTATCAAATGTTATGACTCTTTTGCCAGGAGACATCATTTTGACCGGGACTCCTGGCGGTGTTGGTGTATACAGAAACCCCCCAGAGTTTCTTCAACCCGGTGACGTGATCACGAGTGAAATAGAGAACATTGGTACTTTTGAAACAAGAGTCGAAGAATTTTAA
Protein
MKLLQFSYKNSDGIRAGFLDGDNVVDINKEDPKLPSTLIELLKTNSISKVKKISAKSPVPLSGITLKAPVHGNDKVLCVGLNYKDHCEEQKLTPPELPFIFNKFPSTVVGPNDTIKLKMDVSKAVVCEIELTVVIGKKASKVDSSHAFDYVLGYTIAQDIGATDWEKNKKISQLLLGKAMDTFCPIGPWIVTSDEIGNPQNLNVKCSINGVQKQKSNTNQFIHKIPDIIARLSNVMTLLPGDIILTGTPGGVGVYRNPPEFLQPGDVITSEIENIGTFETRVEEF
Summary
Description
May have hydrolase activity.
Cofactor
Ca(2+)
Mg(2+)
Mg(2+)
Similarity
Belongs to the FAH family.
Keywords
Calcium
Hydrolase
Magnesium
Metal-binding
Acetylation
Complete proteome
Reference proteome
Feature
chain Fumarylacetoacetate hydrolase domain-containing protein 2
Uniprot
H9IUY8
A0A2A4JGP7
A0A2H1VWM9
A0A2W1BQW5
A0A0L7L322
S4PCS0
+ More
A0A3S2NJT7 H9IS36 Q2F601 A0A2H1VFY1 A0A2A4K9I9 A0A2A4K945 A0A2H1V0L4 I4DP61 A0A194QDS5 Q17N03 A0A1B0CS49 A0A023ERA0 A0A1L8E2L4 A0A1L8E2P0 K1QP24 A0A1B6LC38 A0A1L8E9V2 A0A1B6GRW8 B0XIL6 A0A1B6IE47 A0A336L0T5 A0A336KQN9 A0A0A9XLH6 A0A067R380 Q5TMH4 A0A1B6CPP5 A0A182HN30 A0A182VNE1 A0A250YF70 A0A146LA26 A0A1I8PWU6 A0A182LNV2 A0A182WX74 A0A182U545 A0A210PYB4 A0A182KBL7 A0A1Q3FKJ5 A0A1I8MQA1 A0A182YNI4 A0A182PDB0 J9JPT5 A0A182MSH9 A0A182QB46 L8Y998 A0A182MXL1 A0A1Q3FIU4 A0A1Q3FKS1 Q6GLT8 T1DGH7 G1TPZ3 A0A182WEL4 J3JX48 W5PXA6 G1Q7R9 S7MKS3 A0A1S3FGH8 A0A2S2PCF2 V9KVI1 A0A2M3ZAH2 A0A1S3FF86 A0A1S3FEX5 I3NDW8 W5JD71 A0A2M4AM47 E0VV30 A0A1U7Q9F0 A0A3P4PJU0 A0A1L8H1M1 A0A2M4AMK1 A0A2M4AMS7 A0A226N2J6 G3H030 L8IW04 A0A0K8TM05 A0A3Q7VD17 A0A2H8TNV6 M3YCH2 A0A2Y9L439 H3AA15 A0A2Y9HN79 A0A3L7HEG4 A0A2Y9QBB7 A0A402EJ73 T1PDK5 A0A3Q7Q6V4 B2RYW9 A0A384CV90 A0A452R677 A0A3Q7UHT5 A0A0R4IWE1 A0A452FAL7 A0A1A8ESQ4 A0A336KRN5
A0A3S2NJT7 H9IS36 Q2F601 A0A2H1VFY1 A0A2A4K9I9 A0A2A4K945 A0A2H1V0L4 I4DP61 A0A194QDS5 Q17N03 A0A1B0CS49 A0A023ERA0 A0A1L8E2L4 A0A1L8E2P0 K1QP24 A0A1B6LC38 A0A1L8E9V2 A0A1B6GRW8 B0XIL6 A0A1B6IE47 A0A336L0T5 A0A336KQN9 A0A0A9XLH6 A0A067R380 Q5TMH4 A0A1B6CPP5 A0A182HN30 A0A182VNE1 A0A250YF70 A0A146LA26 A0A1I8PWU6 A0A182LNV2 A0A182WX74 A0A182U545 A0A210PYB4 A0A182KBL7 A0A1Q3FKJ5 A0A1I8MQA1 A0A182YNI4 A0A182PDB0 J9JPT5 A0A182MSH9 A0A182QB46 L8Y998 A0A182MXL1 A0A1Q3FIU4 A0A1Q3FKS1 Q6GLT8 T1DGH7 G1TPZ3 A0A182WEL4 J3JX48 W5PXA6 G1Q7R9 S7MKS3 A0A1S3FGH8 A0A2S2PCF2 V9KVI1 A0A2M3ZAH2 A0A1S3FF86 A0A1S3FEX5 I3NDW8 W5JD71 A0A2M4AM47 E0VV30 A0A1U7Q9F0 A0A3P4PJU0 A0A1L8H1M1 A0A2M4AMK1 A0A2M4AMS7 A0A226N2J6 G3H030 L8IW04 A0A0K8TM05 A0A3Q7VD17 A0A2H8TNV6 M3YCH2 A0A2Y9L439 H3AA15 A0A2Y9HN79 A0A3L7HEG4 A0A2Y9QBB7 A0A402EJ73 T1PDK5 A0A3Q7Q6V4 B2RYW9 A0A384CV90 A0A452R677 A0A3Q7UHT5 A0A0R4IWE1 A0A452FAL7 A0A1A8ESQ4 A0A336KRN5
EC Number
3.-.-.-
Pubmed
19121390
28756777
26227816
23622113
22651552
26354079
+ More
17510324 24945155 22992520 25401762 24845553 12364791 14747013 17210077 28087693 26823975 20966253 28812685 25315136 25244985 23385571 21993624 22516182 20809919 24402279 20920257 23761445 20566863 27762356 21804562 22751099 26369729 9215903 29704459 15489334 23594743
17510324 24945155 22992520 25401762 24845553 12364791 14747013 17210077 28087693 26823975 20966253 28812685 25315136 25244985 23385571 21993624 22516182 20809919 24402279 20920257 23761445 20566863 27762356 21804562 22751099 26369729 9215903 29704459 15489334 23594743
EMBL
BABH01000070
NWSH01001435
PCG71237.1
ODYU01004895
SOQ45239.1
KZ149928
+ More
PZC77479.1 JTDY01003234 KOB69908.1 GAIX01004016 JAA88544.1 RSAL01000019 RVE52794.1 BABH01035686 DQ311271 ABD36216.1 ODYU01002352 SOQ39735.1 NWSH01000019 PCG80729.1 PCG80727.1 ODYU01000131 SOQ34390.1 AK403436 BAM19701.1 KQ459144 KPJ03612.1 CH477202 EAT48142.1 AJWK01025642 GAPW01002414 JAC11184.1 GFDF01001104 JAV12980.1 GFDF01001103 JAV12981.1 JH816002 EKC38637.1 GEBQ01018730 JAT21247.1 GFDG01003417 JAV15382.1 GECZ01004610 JAS65159.1 DS233329 EDS29440.1 GECU01022497 JAS85209.1 UFQS01000773 UFQT01000773 SSX06780.1 SSX27125.1 SSX06779.1 SSX27124.1 GBHO01022780 GBHO01022778 GBRD01006623 GBRD01006622 JAG20824.1 JAG20826.1 JAG59198.1 KK852941 KDR13529.1 AAAB01008986 EAL38923.3 GEDC01021841 JAS15457.1 APCN01001831 GFFW01002505 JAV42283.1 GDHC01014194 JAQ04435.1 NEDP02005386 OWF41480.1 GFDL01007013 JAV28032.1 ABLF02030307 AXCM01002192 AXCN02001066 KB365080 ELV11525.1 GFDL01007642 JAV27403.1 GFDL01006969 JAV28076.1 BC074365 GAMD01002771 JAA98819.1 AAGW02007409 BT127816 AEE62778.1 AMGL01078866 AAPE02041650 KE161478 EPQ04105.1 GGMR01014456 MBY27075.1 JW870532 AFP03050.1 GGFM01004724 MBW25475.1 AGTP01102139 ADMH02001501 ETN62292.1 GGFK01008523 MBW41844.1 DS235802 EEB17236.1 CYRY02032846 VCX10250.1 CM004470 OCT89956.1 GGFK01008656 MBW41977.1 GGFK01008587 MBW41908.1 MCFN01000251 OXB61816.1 JH000086 EGW04973.1 JH880729 ELR59332.1 GDAI01002214 JAI15389.1 GFXV01004058 MBW15863.1 AEYP01076409 AFYH01183888 RAZU01000234 RLQ64354.1 BDOT01000015 GCF44253.1 KA645988 KA649281 AFP60617.1 BC166933 CU683877 FP101906 LWLT01000009 HAEB01003648 SBQ50175.1 SSX06778.1 SSX27123.1
PZC77479.1 JTDY01003234 KOB69908.1 GAIX01004016 JAA88544.1 RSAL01000019 RVE52794.1 BABH01035686 DQ311271 ABD36216.1 ODYU01002352 SOQ39735.1 NWSH01000019 PCG80729.1 PCG80727.1 ODYU01000131 SOQ34390.1 AK403436 BAM19701.1 KQ459144 KPJ03612.1 CH477202 EAT48142.1 AJWK01025642 GAPW01002414 JAC11184.1 GFDF01001104 JAV12980.1 GFDF01001103 JAV12981.1 JH816002 EKC38637.1 GEBQ01018730 JAT21247.1 GFDG01003417 JAV15382.1 GECZ01004610 JAS65159.1 DS233329 EDS29440.1 GECU01022497 JAS85209.1 UFQS01000773 UFQT01000773 SSX06780.1 SSX27125.1 SSX06779.1 SSX27124.1 GBHO01022780 GBHO01022778 GBRD01006623 GBRD01006622 JAG20824.1 JAG20826.1 JAG59198.1 KK852941 KDR13529.1 AAAB01008986 EAL38923.3 GEDC01021841 JAS15457.1 APCN01001831 GFFW01002505 JAV42283.1 GDHC01014194 JAQ04435.1 NEDP02005386 OWF41480.1 GFDL01007013 JAV28032.1 ABLF02030307 AXCM01002192 AXCN02001066 KB365080 ELV11525.1 GFDL01007642 JAV27403.1 GFDL01006969 JAV28076.1 BC074365 GAMD01002771 JAA98819.1 AAGW02007409 BT127816 AEE62778.1 AMGL01078866 AAPE02041650 KE161478 EPQ04105.1 GGMR01014456 MBY27075.1 JW870532 AFP03050.1 GGFM01004724 MBW25475.1 AGTP01102139 ADMH02001501 ETN62292.1 GGFK01008523 MBW41844.1 DS235802 EEB17236.1 CYRY02032846 VCX10250.1 CM004470 OCT89956.1 GGFK01008656 MBW41977.1 GGFK01008587 MBW41908.1 MCFN01000251 OXB61816.1 JH000086 EGW04973.1 JH880729 ELR59332.1 GDAI01002214 JAI15389.1 GFXV01004058 MBW15863.1 AEYP01076409 AFYH01183888 RAZU01000234 RLQ64354.1 BDOT01000015 GCF44253.1 KA645988 KA649281 AFP60617.1 BC166933 CU683877 FP101906 LWLT01000009 HAEB01003648 SBQ50175.1 SSX06778.1 SSX27123.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053268
UP000008820
+ More
UP000092461 UP000005408 UP000002320 UP000027135 UP000007062 UP000075840 UP000075903 UP000095300 UP000075882 UP000076407 UP000075902 UP000242188 UP000075881 UP000095301 UP000076408 UP000075885 UP000007819 UP000075883 UP000075886 UP000011518 UP000075884 UP000001811 UP000075920 UP000002356 UP000001074 UP000081671 UP000005215 UP000000673 UP000009046 UP000189706 UP000186698 UP000198323 UP000001075 UP000286642 UP000000715 UP000248482 UP000008672 UP000248481 UP000273346 UP000248483 UP000288954 UP000286641 UP000002494 UP000261680 UP000291022 UP000000437 UP000291000
UP000092461 UP000005408 UP000002320 UP000027135 UP000007062 UP000075840 UP000075903 UP000095300 UP000075882 UP000076407 UP000075902 UP000242188 UP000075881 UP000095301 UP000076408 UP000075885 UP000007819 UP000075883 UP000075886 UP000011518 UP000075884 UP000001811 UP000075920 UP000002356 UP000001074 UP000081671 UP000005215 UP000000673 UP000009046 UP000189706 UP000186698 UP000198323 UP000001075 UP000286642 UP000000715 UP000248482 UP000008672 UP000248481 UP000273346 UP000248483 UP000288954 UP000286641 UP000002494 UP000261680 UP000291022 UP000000437 UP000291000
Interpro
Gene 3D
ProteinModelPortal
H9IUY8
A0A2A4JGP7
A0A2H1VWM9
A0A2W1BQW5
A0A0L7L322
S4PCS0
+ More
A0A3S2NJT7 H9IS36 Q2F601 A0A2H1VFY1 A0A2A4K9I9 A0A2A4K945 A0A2H1V0L4 I4DP61 A0A194QDS5 Q17N03 A0A1B0CS49 A0A023ERA0 A0A1L8E2L4 A0A1L8E2P0 K1QP24 A0A1B6LC38 A0A1L8E9V2 A0A1B6GRW8 B0XIL6 A0A1B6IE47 A0A336L0T5 A0A336KQN9 A0A0A9XLH6 A0A067R380 Q5TMH4 A0A1B6CPP5 A0A182HN30 A0A182VNE1 A0A250YF70 A0A146LA26 A0A1I8PWU6 A0A182LNV2 A0A182WX74 A0A182U545 A0A210PYB4 A0A182KBL7 A0A1Q3FKJ5 A0A1I8MQA1 A0A182YNI4 A0A182PDB0 J9JPT5 A0A182MSH9 A0A182QB46 L8Y998 A0A182MXL1 A0A1Q3FIU4 A0A1Q3FKS1 Q6GLT8 T1DGH7 G1TPZ3 A0A182WEL4 J3JX48 W5PXA6 G1Q7R9 S7MKS3 A0A1S3FGH8 A0A2S2PCF2 V9KVI1 A0A2M3ZAH2 A0A1S3FF86 A0A1S3FEX5 I3NDW8 W5JD71 A0A2M4AM47 E0VV30 A0A1U7Q9F0 A0A3P4PJU0 A0A1L8H1M1 A0A2M4AMK1 A0A2M4AMS7 A0A226N2J6 G3H030 L8IW04 A0A0K8TM05 A0A3Q7VD17 A0A2H8TNV6 M3YCH2 A0A2Y9L439 H3AA15 A0A2Y9HN79 A0A3L7HEG4 A0A2Y9QBB7 A0A402EJ73 T1PDK5 A0A3Q7Q6V4 B2RYW9 A0A384CV90 A0A452R677 A0A3Q7UHT5 A0A0R4IWE1 A0A452FAL7 A0A1A8ESQ4 A0A336KRN5
A0A3S2NJT7 H9IS36 Q2F601 A0A2H1VFY1 A0A2A4K9I9 A0A2A4K945 A0A2H1V0L4 I4DP61 A0A194QDS5 Q17N03 A0A1B0CS49 A0A023ERA0 A0A1L8E2L4 A0A1L8E2P0 K1QP24 A0A1B6LC38 A0A1L8E9V2 A0A1B6GRW8 B0XIL6 A0A1B6IE47 A0A336L0T5 A0A336KQN9 A0A0A9XLH6 A0A067R380 Q5TMH4 A0A1B6CPP5 A0A182HN30 A0A182VNE1 A0A250YF70 A0A146LA26 A0A1I8PWU6 A0A182LNV2 A0A182WX74 A0A182U545 A0A210PYB4 A0A182KBL7 A0A1Q3FKJ5 A0A1I8MQA1 A0A182YNI4 A0A182PDB0 J9JPT5 A0A182MSH9 A0A182QB46 L8Y998 A0A182MXL1 A0A1Q3FIU4 A0A1Q3FKS1 Q6GLT8 T1DGH7 G1TPZ3 A0A182WEL4 J3JX48 W5PXA6 G1Q7R9 S7MKS3 A0A1S3FGH8 A0A2S2PCF2 V9KVI1 A0A2M3ZAH2 A0A1S3FF86 A0A1S3FEX5 I3NDW8 W5JD71 A0A2M4AM47 E0VV30 A0A1U7Q9F0 A0A3P4PJU0 A0A1L8H1M1 A0A2M4AMK1 A0A2M4AMS7 A0A226N2J6 G3H030 L8IW04 A0A0K8TM05 A0A3Q7VD17 A0A2H8TNV6 M3YCH2 A0A2Y9L439 H3AA15 A0A2Y9HN79 A0A3L7HEG4 A0A2Y9QBB7 A0A402EJ73 T1PDK5 A0A3Q7Q6V4 B2RYW9 A0A384CV90 A0A452R677 A0A3Q7UHT5 A0A0R4IWE1 A0A452FAL7 A0A1A8ESQ4 A0A336KRN5
PDB
3QDF
E-value=5.00593e-37,
Score=386
Ontologies
KEGG
GO
PANTHER
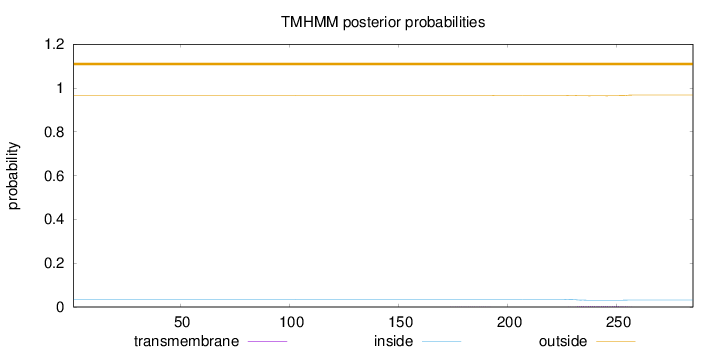
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09916
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03478
outside
1 - 285
Population Genetic Test Statistics
Pi
11.56217
Theta
17.197664
Tajima's D
-0.915879
CLR
16.149706
CSRT
0.152842357882106
Interpretation
Uncertain
