Gene
KWMTBOMO08117
Pre Gene Modal
BGIBMGA001064
Annotation
PREDICTED:_polypeptide_N-acetylgalactosaminyltransferase_10-like_[Bombyx_mori]
Full name
Polypeptide N-acetylgalactosaminyltransferase
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
Location in the cell
Cytoplasmic Reliability : 1.701 Mitochondrial Reliability : 1.26
Sequence
CDS
ATGATGTTGCGACGCAGTTTGTTTCGTTTATTGAGGCTGCGAAATCTTGTATTTGTAACGTTACTAGTCATCGTGTTATATTTAGTAAAGTCATTGAGTTATTCTGAGTCTGATAAGCTAATTGTTAGAAGAGAAGCTTTAGCTCACTCAGATGAGCTGATGAACCATGATACTGCTGTACTACATGCTCCACCCGATTTGAACAAGGCTGTTAATAAAGAGAACCATTTTGTACCAAAGAGTTTGAAGAAAATAGACTGGCATAACTATGAACAGATAAGCAGTGAGAGGAAACGTGTGGGTATTGGTGAACAAGGTCTCCCAGCGCATTTACCAAAAGAAGAATCAGATTTAGAAGAATCATTGTATTCAGTGAATGGTTTCAATGGTGCTCTAAGTGATAAAATACCATTGAGCCGTTCATTGCCAGACATAAGACATTCAGGATGTAAAAAAAAAATGTATATCGAATCTTTGCCGAGCGTCAGTGTGGTTGTACCGTTTCACAATGAGCACTGGAGTACGTTGTTGAGGACAGCATATAGCGTGCTGTACAGATCACCAGAGCATCTCATTAAGGAGATATTTTTGGTGGATGATGCTAGTACCAAAGATTTTCTAGGGAAAGCATTAGATGACTACCTAGAGGGAAACATGCCTAAAGTGAAAGTGATCAGACTGCCGAAACGCAGTGGACTGATCACGGCTCGGCTGGCGGGCGCAGAGCGAGCGACGGCCGACGTCCTGATATTCCTGGACTCGCACACGGAGGCCAACGTCAACTGGCTCCCGCCCCTGCTAGAACCCATAGCCCTGGACTACCGGACCGTGATGTGTCCCTTCATTGACGTGATAGCGTTCGATACCTTCGAGTACAGGGCTCAGGACGAAGGTGCCAGGGGAGCTTTCGATTGGGAGTTCTTCTACAAAAGGCTTCCCGTCCTCCCTAAGGACGAAAAGAATATGCCCGAGCCCTTCGAGAGCCCCGTGATGGCGGGCGGTCTGTTCGCGATATCGCGTGCATTCTTCTGGGAGCTGGGCGGTTACGACCCGGGCCTCGACATATGGGGCGGGGAACAATACGAACTCAGCTTTAAGATATGGCAATGCGGAGGTCGGATGCTGGACGCGCCGTGCTCCCGAGTGGGTCACATTTACCGGAAGTTCGCCCCGTTCCCGAACCCCGGGCACGGAGACTTCGTGGGAAAGAACTACCGGCGGGTGGCCGAGGTGTGGATGGACGAGTACGCGGAGCACCTGTACCGGCGCCGCCCGCACTACCGCCGCATCGACCCCGGGGACCTCGCGCCGCAGAAGCGCCTCCGGGAGAAGCTCAACTGTAAGCCCTTCAAGTGGTTCATGACTCAGATAGCGTTTGATTTGACGGCGAAGTACCCGCCCGTGGAGCCTAAGCCTTTTGCCGAGGGAAAGGTAAGAGATTCGTTTATTTCAACTGTCTTGAAAACTATTGATTGTCATTGCTTAGTAAGGTACTGTAGTCTGTTAATAAGTAGTCAATAA
Protein
MMLRRSLFRLLRLRNLVFVTLLVIVLYLVKSLSYSESDKLIVRREALAHSDELMNHDTAVLHAPPDLNKAVNKENHFVPKSLKKIDWHNYEQISSERKRVGIGEQGLPAHLPKEESDLEESLYSVNGFNGALSDKIPLSRSLPDIRHSGCKKKMYIESLPSVSVVVPFHNEHWSTLLRTAYSVLYRSPEHLIKEIFLVDDASTKDFLGKALDDYLEGNMPKVKVIRLPKRSGLITARLAGAERATADVLIFLDSHTEANVNWLPPLLEPIALDYRTVMCPFIDVIAFDTFEYRAQDEGARGAFDWEFFYKRLPVLPKDEKNMPEPFESPVMAGGLFAISRAFFWELGGYDPGLDIWGGEQYELSFKIWQCGGRMLDAPCSRVGHIYRKFAPFPNPGHGDFVGKNYRRVAEVWMDEYAEHLYRRRPHYRRIDPGDLAPQKRLREKLNCKPFKWFMTQIAFDLTAKYPPVEPKPFAEGKVRDSFISTVLKTIDCHCLVRYCSLLISSQ
Summary
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Belongs to the cyclin family.
Belongs to the cyclin family.
Feature
chain Polypeptide N-acetylgalactosaminyltransferase
Uniprot
H9IUY4
A0A2A4K2U5
A0A194RFJ9
A0A2H1WSN0
A0A194Q686
E0VB90
+ More
V5I8X7 D2A164 A0A1Y1NDW2 A0A1B6HML5 A0A2J7PXF1 A0A2J7PXG0 A0A2J7PXF8 A0A1B6ED34 A0A182MLB1 A0A182WEW8 A0A182FBB8 A0A182QKA4 A0A182JG65 A0A182Y1G1 U4TVW9 N6UJQ0 A0A084WIB1 A0A2M4BHE1 A0A182XR66 A0A1S4H976 A0A1C7CZK7 A0A232F8X5 A0A182IFR1 A0A2M4BIF4 Q7Q0E9 A0A182UPG0 T1JNW7 A0A2M4AE52 A0A2M4AE47 A0A1W0WCD2 A0A2M3Z2T2 A0A1S4EWG7 A0A3R7PH79 A0A1Q3FAN3 A0A1Q3FAI0 W5JKD2 Q17NN5 A0A2A3EC73 A0A088A602 A0A026WBU0 B0X3N7 B0WFR4 A0A067QGM9 A0A182GFJ3 A0A182HBH2 E9J1W6 A0A1S3JMP6 A0A158NJ53 A0A0L7QMB6 A0A2H8TMN6 A0A195BJU4 A0A151JRT7 F4W772 J9K6S1 A0A2M3Z5C4 A0A2T7PMQ0 A0A195EYQ3 A0A154P465 R7V610 A0A210PT88 A0A147BRJ6 E2A417 A0A1B6CII9 A0A0J7KFZ5 A0A195C355 A0A2M3Z5M7 A0A151WV38 A0A1B0DP26 U5ERV4 V4ALX2 A0A293LYI6 A0A1Z5LFV8 A0A1E1XJW7 A0A1E1X6U5 L7M0Z7 A0A0P4VZA6 A0A0P4VSE9 A0A1J1J6K8 A0A0P6H812 A0A1L8E1A6 A0A0P4YXZ7 A0A267EH10 A0A0N8BQ37 A0A0P5VDW3 A0A0P5E8P0 K1PQC0 A0A2P2I4L2 A0A0P5BA74 A0A1I8GGZ3 A0A0P5X1F8 E9HP56 A0A0P5B2H8 A0A2P8ZAR9
V5I8X7 D2A164 A0A1Y1NDW2 A0A1B6HML5 A0A2J7PXF1 A0A2J7PXG0 A0A2J7PXF8 A0A1B6ED34 A0A182MLB1 A0A182WEW8 A0A182FBB8 A0A182QKA4 A0A182JG65 A0A182Y1G1 U4TVW9 N6UJQ0 A0A084WIB1 A0A2M4BHE1 A0A182XR66 A0A1S4H976 A0A1C7CZK7 A0A232F8X5 A0A182IFR1 A0A2M4BIF4 Q7Q0E9 A0A182UPG0 T1JNW7 A0A2M4AE52 A0A2M4AE47 A0A1W0WCD2 A0A2M3Z2T2 A0A1S4EWG7 A0A3R7PH79 A0A1Q3FAN3 A0A1Q3FAI0 W5JKD2 Q17NN5 A0A2A3EC73 A0A088A602 A0A026WBU0 B0X3N7 B0WFR4 A0A067QGM9 A0A182GFJ3 A0A182HBH2 E9J1W6 A0A1S3JMP6 A0A158NJ53 A0A0L7QMB6 A0A2H8TMN6 A0A195BJU4 A0A151JRT7 F4W772 J9K6S1 A0A2M3Z5C4 A0A2T7PMQ0 A0A195EYQ3 A0A154P465 R7V610 A0A210PT88 A0A147BRJ6 E2A417 A0A1B6CII9 A0A0J7KFZ5 A0A195C355 A0A2M3Z5M7 A0A151WV38 A0A1B0DP26 U5ERV4 V4ALX2 A0A293LYI6 A0A1Z5LFV8 A0A1E1XJW7 A0A1E1X6U5 L7M0Z7 A0A0P4VZA6 A0A0P4VSE9 A0A1J1J6K8 A0A0P6H812 A0A1L8E1A6 A0A0P4YXZ7 A0A267EH10 A0A0N8BQ37 A0A0P5VDW3 A0A0P5E8P0 K1PQC0 A0A2P2I4L2 A0A0P5BA74 A0A1I8GGZ3 A0A0P5X1F8 E9HP56 A0A0P5B2H8 A0A2P8ZAR9
EC Number
2.4.1.-
Pubmed
19121390
26354079
20566863
18362917
19820115
28004739
+ More
25244985 23537049 24438588 12364791 20966253 28648823 20920257 23761445 17510324 24508170 30249741 24845553 26483478 21282665 21347285 21719571 23254933 28812685 29652888 20798317 28528879 29209593 28503490 25576852 22992520 26392545 21292972 29403074
25244985 23537049 24438588 12364791 20966253 28648823 20920257 23761445 17510324 24508170 30249741 24845553 26483478 21282665 21347285 21719571 23254933 28812685 29652888 20798317 28528879 29209593 28503490 25576852 22992520 26392545 21292972 29403074
EMBL
BABH01000157
NWSH01000210
PCG78369.1
KQ460297
KPJ16234.1
ODYU01010728
+ More
SOQ56017.1 KQ459439 KPJ01048.1 DS235023 EEB10646.1 GALX01003915 JAB64551.1 KQ971338 EFA02618.2 GEZM01005562 JAV96093.1 GECU01031820 JAS75886.1 NEVH01020859 PNF21012.1 PNF21010.1 PNF21009.1 GEDC01001455 JAS35843.1 AXCM01001414 AXCN02001027 KB631743 ERL85724.1 APGK01021331 KB740314 ENN80896.1 ATLV01023925 KE525347 KFB49955.1 GGFJ01003316 MBW52457.1 AAAB01008986 NNAY01000700 OXU26918.1 APCN01003311 GGFJ01003653 MBW52794.1 EAA00190.4 JH432064 GGFK01005743 MBW39064.1 GGFK01005744 MBW39065.1 MTYJ01000136 OQV12822.1 GGFM01002075 MBW22826.1 QCYY01002268 ROT71654.1 GFDL01010473 JAV24572.1 GFDL01010523 JAV24522.1 ADMH02001279 ETN63234.1 CH477198 EAT48278.1 KZ288305 PBC28776.1 KK107293 QOIP01000005 EZA53438.1 RLU22425.1 DS232318 EDS39929.1 DS231920 EDS26408.1 KK853499 KDR07167.1 JXUM01059638 KQ562064 KXJ76761.1 JXUM01124915 KQ566885 KXJ69756.1 GL767674 EFZ13283.1 ADTU01017493 KQ414894 KOC59762.1 GFXV01003117 MBW14922.1 KQ976453 KYM85443.1 KQ978579 KYN30004.1 GL887813 EGI69956.1 ABLF02039966 GGFM01002954 MBW23705.1 PZQS01000003 PVD34693.1 KQ981920 KYN33014.1 KQ434809 KZC06633.1 AMQN01004963 KB294813 ELU14019.1 NEDP02005513 OWF39703.1 GEGO01002016 JAR93388.1 GL436519 EFN71869.1 GEDC01024077 JAS13221.1 LBMM01008031 KMQ89202.1 KQ978317 KYM95272.1 GGFM01003054 MBW23805.1 KQ982708 KYQ51789.1 AJVK01007905 AJVK01007906 GANO01003546 JAB56325.1 KB201271 ESO98122.1 GFWV01009044 MAA33773.1 GFJQ02000687 JAW06283.1 GFAA01003869 JAT99565.1 GFAC01004203 JAT94985.1 GACK01007319 JAA57715.1 GDRN01101392 JAI58386.1 GDRN01101393 JAI58385.1 CVRI01000067 CRL06521.1 GDIQ01023411 JAN71326.1 GFDF01001574 JAV12510.1 GDIP01221378 JAJ02024.1 NIVC01002109 PAA60805.1 GDIQ01155963 JAK95762.1 GDIP01101730 JAM01985.1 GDIP01151095 JAJ72307.1 JH818840 EKC18555.1 IACF01003356 LAB68973.1 GDIP01188134 JAJ35268.1 NIVC01000861 PAA75794.1 GDIP01079100 JAM24615.1 GL732703 EFX66478.1 GDIP01190726 JAJ32676.1 PYGN01000121 PSN53588.1
SOQ56017.1 KQ459439 KPJ01048.1 DS235023 EEB10646.1 GALX01003915 JAB64551.1 KQ971338 EFA02618.2 GEZM01005562 JAV96093.1 GECU01031820 JAS75886.1 NEVH01020859 PNF21012.1 PNF21010.1 PNF21009.1 GEDC01001455 JAS35843.1 AXCM01001414 AXCN02001027 KB631743 ERL85724.1 APGK01021331 KB740314 ENN80896.1 ATLV01023925 KE525347 KFB49955.1 GGFJ01003316 MBW52457.1 AAAB01008986 NNAY01000700 OXU26918.1 APCN01003311 GGFJ01003653 MBW52794.1 EAA00190.4 JH432064 GGFK01005743 MBW39064.1 GGFK01005744 MBW39065.1 MTYJ01000136 OQV12822.1 GGFM01002075 MBW22826.1 QCYY01002268 ROT71654.1 GFDL01010473 JAV24572.1 GFDL01010523 JAV24522.1 ADMH02001279 ETN63234.1 CH477198 EAT48278.1 KZ288305 PBC28776.1 KK107293 QOIP01000005 EZA53438.1 RLU22425.1 DS232318 EDS39929.1 DS231920 EDS26408.1 KK853499 KDR07167.1 JXUM01059638 KQ562064 KXJ76761.1 JXUM01124915 KQ566885 KXJ69756.1 GL767674 EFZ13283.1 ADTU01017493 KQ414894 KOC59762.1 GFXV01003117 MBW14922.1 KQ976453 KYM85443.1 KQ978579 KYN30004.1 GL887813 EGI69956.1 ABLF02039966 GGFM01002954 MBW23705.1 PZQS01000003 PVD34693.1 KQ981920 KYN33014.1 KQ434809 KZC06633.1 AMQN01004963 KB294813 ELU14019.1 NEDP02005513 OWF39703.1 GEGO01002016 JAR93388.1 GL436519 EFN71869.1 GEDC01024077 JAS13221.1 LBMM01008031 KMQ89202.1 KQ978317 KYM95272.1 GGFM01003054 MBW23805.1 KQ982708 KYQ51789.1 AJVK01007905 AJVK01007906 GANO01003546 JAB56325.1 KB201271 ESO98122.1 GFWV01009044 MAA33773.1 GFJQ02000687 JAW06283.1 GFAA01003869 JAT99565.1 GFAC01004203 JAT94985.1 GACK01007319 JAA57715.1 GDRN01101392 JAI58386.1 GDRN01101393 JAI58385.1 CVRI01000067 CRL06521.1 GDIQ01023411 JAN71326.1 GFDF01001574 JAV12510.1 GDIP01221378 JAJ02024.1 NIVC01002109 PAA60805.1 GDIQ01155963 JAK95762.1 GDIP01101730 JAM01985.1 GDIP01151095 JAJ72307.1 JH818840 EKC18555.1 IACF01003356 LAB68973.1 GDIP01188134 JAJ35268.1 NIVC01000861 PAA75794.1 GDIP01079100 JAM24615.1 GL732703 EFX66478.1 GDIP01190726 JAJ32676.1 PYGN01000121 PSN53588.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000009046
UP000007266
+ More
UP000235965 UP000075883 UP000075920 UP000069272 UP000075886 UP000075880 UP000076408 UP000030742 UP000019118 UP000030765 UP000076407 UP000075882 UP000215335 UP000075840 UP000007062 UP000075903 UP000283509 UP000000673 UP000008820 UP000242457 UP000005203 UP000053097 UP000279307 UP000002320 UP000027135 UP000069940 UP000249989 UP000085678 UP000005205 UP000053825 UP000078540 UP000078492 UP000007755 UP000007819 UP000245119 UP000078541 UP000076502 UP000014760 UP000242188 UP000000311 UP000036403 UP000078542 UP000075809 UP000092462 UP000030746 UP000183832 UP000215902 UP000005408 UP000095280 UP000000305 UP000245037
UP000235965 UP000075883 UP000075920 UP000069272 UP000075886 UP000075880 UP000076408 UP000030742 UP000019118 UP000030765 UP000076407 UP000075882 UP000215335 UP000075840 UP000007062 UP000075903 UP000283509 UP000000673 UP000008820 UP000242457 UP000005203 UP000053097 UP000279307 UP000002320 UP000027135 UP000069940 UP000249989 UP000085678 UP000005205 UP000053825 UP000078540 UP000078492 UP000007755 UP000007819 UP000245119 UP000078541 UP000076502 UP000014760 UP000242188 UP000000311 UP000036403 UP000078542 UP000075809 UP000092462 UP000030746 UP000183832 UP000215902 UP000005408 UP000095280 UP000000305 UP000245037
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9IUY4
A0A2A4K2U5
A0A194RFJ9
A0A2H1WSN0
A0A194Q686
E0VB90
+ More
V5I8X7 D2A164 A0A1Y1NDW2 A0A1B6HML5 A0A2J7PXF1 A0A2J7PXG0 A0A2J7PXF8 A0A1B6ED34 A0A182MLB1 A0A182WEW8 A0A182FBB8 A0A182QKA4 A0A182JG65 A0A182Y1G1 U4TVW9 N6UJQ0 A0A084WIB1 A0A2M4BHE1 A0A182XR66 A0A1S4H976 A0A1C7CZK7 A0A232F8X5 A0A182IFR1 A0A2M4BIF4 Q7Q0E9 A0A182UPG0 T1JNW7 A0A2M4AE52 A0A2M4AE47 A0A1W0WCD2 A0A2M3Z2T2 A0A1S4EWG7 A0A3R7PH79 A0A1Q3FAN3 A0A1Q3FAI0 W5JKD2 Q17NN5 A0A2A3EC73 A0A088A602 A0A026WBU0 B0X3N7 B0WFR4 A0A067QGM9 A0A182GFJ3 A0A182HBH2 E9J1W6 A0A1S3JMP6 A0A158NJ53 A0A0L7QMB6 A0A2H8TMN6 A0A195BJU4 A0A151JRT7 F4W772 J9K6S1 A0A2M3Z5C4 A0A2T7PMQ0 A0A195EYQ3 A0A154P465 R7V610 A0A210PT88 A0A147BRJ6 E2A417 A0A1B6CII9 A0A0J7KFZ5 A0A195C355 A0A2M3Z5M7 A0A151WV38 A0A1B0DP26 U5ERV4 V4ALX2 A0A293LYI6 A0A1Z5LFV8 A0A1E1XJW7 A0A1E1X6U5 L7M0Z7 A0A0P4VZA6 A0A0P4VSE9 A0A1J1J6K8 A0A0P6H812 A0A1L8E1A6 A0A0P4YXZ7 A0A267EH10 A0A0N8BQ37 A0A0P5VDW3 A0A0P5E8P0 K1PQC0 A0A2P2I4L2 A0A0P5BA74 A0A1I8GGZ3 A0A0P5X1F8 E9HP56 A0A0P5B2H8 A0A2P8ZAR9
V5I8X7 D2A164 A0A1Y1NDW2 A0A1B6HML5 A0A2J7PXF1 A0A2J7PXG0 A0A2J7PXF8 A0A1B6ED34 A0A182MLB1 A0A182WEW8 A0A182FBB8 A0A182QKA4 A0A182JG65 A0A182Y1G1 U4TVW9 N6UJQ0 A0A084WIB1 A0A2M4BHE1 A0A182XR66 A0A1S4H976 A0A1C7CZK7 A0A232F8X5 A0A182IFR1 A0A2M4BIF4 Q7Q0E9 A0A182UPG0 T1JNW7 A0A2M4AE52 A0A2M4AE47 A0A1W0WCD2 A0A2M3Z2T2 A0A1S4EWG7 A0A3R7PH79 A0A1Q3FAN3 A0A1Q3FAI0 W5JKD2 Q17NN5 A0A2A3EC73 A0A088A602 A0A026WBU0 B0X3N7 B0WFR4 A0A067QGM9 A0A182GFJ3 A0A182HBH2 E9J1W6 A0A1S3JMP6 A0A158NJ53 A0A0L7QMB6 A0A2H8TMN6 A0A195BJU4 A0A151JRT7 F4W772 J9K6S1 A0A2M3Z5C4 A0A2T7PMQ0 A0A195EYQ3 A0A154P465 R7V610 A0A210PT88 A0A147BRJ6 E2A417 A0A1B6CII9 A0A0J7KFZ5 A0A195C355 A0A2M3Z5M7 A0A151WV38 A0A1B0DP26 U5ERV4 V4ALX2 A0A293LYI6 A0A1Z5LFV8 A0A1E1XJW7 A0A1E1X6U5 L7M0Z7 A0A0P4VZA6 A0A0P4VSE9 A0A1J1J6K8 A0A0P6H812 A0A1L8E1A6 A0A0P4YXZ7 A0A267EH10 A0A0N8BQ37 A0A0P5VDW3 A0A0P5E8P0 K1PQC0 A0A2P2I4L2 A0A0P5BA74 A0A1I8GGZ3 A0A0P5X1F8 E9HP56 A0A0P5B2H8 A0A2P8ZAR9
PDB
2D7R
E-value=1.14588e-139,
Score=1274
Ontologies
PATHWAY
Topology
Subcellular location
Golgi apparatus membrane
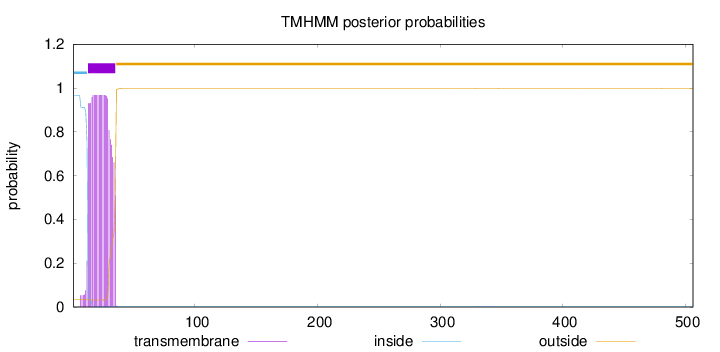
Length:
506
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.9988
Exp number, first 60 AAs:
20.99355
Total prob of N-in:
0.96563
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 506
Population Genetic Test Statistics
Pi
20.926846
Theta
15.301911
Tajima's D
0.518793
CLR
0.842997
CSRT
0.526123693815309
Interpretation
Uncertain
