Gene
KWMTBOMO08116 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001097
Annotation
PREDICTED:_calnexin_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.434 ER Reliability : 1.396 Nuclear Reliability : 1.46
Sequence
CDS
ATGGCTGAAATAAGCCGCAAGTTATTTCTATTTAGTATATTGTTGGCATTTTTCGGCAATACCAGGCTAACAAATGCAAACGAAGTCGACCAAAATGATGAGGCAACTGTGGAGATTGAGGAAGATGATCAATATGTCAGCCCAGAATTGAATTCAGACAATGTGTACCTGTCTGAGCATTTTGATGATGAAAATGCTTTTAAAAGGAAATGGATTAAATCTGAAGCTAAAAAACAGGGAGTAGATGAAAACATAGCGAAATATGACGGAAAATGGGAGATTCAACAACCAATCAGGAAGATCCTAAAGAACGATTTTGGACTTGTCCTCACAACCGAAGCAAAACATGCAGCTATATCTACACTACTGGATAAACCATTTGAGTTTAAAAACAAGCCTCTCATTGTTCAATATGAAGTCACTATGCAGGAAGGACAGAACTGCGGTGGTGCCTACATCAAGCTACTATCAAAAGGGACTAACACGAAAGCCGATCTACGTAAATTCCACGATCAAACTCCGTACACTATAATGTTTGGGCCGGACAAGTGCGGCAATGATAATAAACTACATTTCATATTCAGACATAAGAATCCTAAGAATGGCACCATTGAAGAGAAACACAGTAAGAAACCAACTCAGCGACTAGATGACATCTACAAAGACAAGGAACCTCATTTATACACTCTGATTGTCCGACCAGACAATACCTTCTCTATACTAGTTGACAACAAGGAAGTCAATGCTGGATCTCTGCTAGAGGACTTTACTCCACCTGTCAACCCACCAGAGGAAATTGACGACCCTAATGACAAGAAACCAGAAGATTGGGATGAAAGGGAAAAGATTGTTGATCCTAGTGCTACCAAGCCTGATGACTGGGACGAAACTGAGCCGGCCCAAATTAAAGATCCCAATGCAATTAAACCTGAAGGATGGCTTGACGATGAACCAGAAATGATTCCAGATCCGTCGGCGGTGAAGCCGTCCGACTGGGACGAGGAGATGGACGGCACGTGGGAGGCCGCGCTCGTTGACAACCCTGCGTGCGAGTCGGCGCCGGGCTGCGGCCAGTGGGCGCCCCCCACCGTGCCCAACCCTAACTACAAGGGCATATGGCGAGCTCCTCTGATCCCGAACCCCAACTACAAGGGCAAGTGGAACCCGAGGCGCATTCCGAACCCGGACTACTTCAAGGACGATCACCCCTTCCAGATGACACCCATCCACGGCGTGGGGTTCGAGCTGTGGTCCATGTCCCCGATGCTGCTGTTCGACAACGTGCTGGTTACGGACGACTTCGCGCTGGCGGAGCGCTGGGCGCAGCAGACCTTCGTGCTCAAGAAGGCCAAGCTTTCCAGGGACTCTGATTCGTTTTTGGACCGCGGTCTGAAGTACGCCGGGGACAATCCCTGGGTTTACGCCGTCGTGATCCTCGGAGCCTTCCTGGTCATTGGCTTCGTGACCTATCTCCTGTGCGGCCCTAAATCTGAGCCAGAAGCAGATCCCAAGAAGACAGACGCATTTGTGGAAGACGACCCTCATCAATCGGAAGCCGAAGAGGATCTCAAAGAGGAGGAGCCTCGGGAGAAACCCACCAAGACCAAAGCTGATCTCGAGGGTCCGGAGGAAGAAGTCGAGAACGTAACTTTGACTACGGAGGCGAGCCCAACAGAAACAGAGGGCGCTGGCGACGGACACAGGAAACGGAAGCCTCGTAAGGAGTAA
Protein
MAEISRKLFLFSILLAFFGNTRLTNANEVDQNDEATVEIEEDDQYVSPELNSDNVYLSEHFDDENAFKRKWIKSEAKKQGVDENIAKYDGKWEIQQPIRKILKNDFGLVLTTEAKHAAISTLLDKPFEFKNKPLIVQYEVTMQEGQNCGGAYIKLLSKGTNTKADLRKFHDQTPYTIMFGPDKCGNDNKLHFIFRHKNPKNGTIEEKHSKKPTQRLDDIYKDKEPHLYTLIVRPDNTFSILVDNKEVNAGSLLEDFTPPVNPPEEIDDPNDKKPEDWDEREKIVDPSATKPDDWDETEPAQIKDPNAIKPEGWLDDEPEMIPDPSAVKPSDWDEEMDGTWEAALVDNPACESAPGCGQWAPPTVPNPNYKGIWRAPLIPNPNYKGKWNPRRIPNPDYFKDDHPFQMTPIHGVGFELWSMSPMLLFDNVLVTDDFALAERWAQQTFVLKKAKLSRDSDSFLDRGLKYAGDNPWVYAVVILGAFLVIGFVTYLLCGPKSEPEADPKKTDAFVEDDPHQSEAEEDLKEEEPREKPTKTKADLEGPEEEVENVTLTTEASPTETEGAGDGHRKRKPRKE
Summary
Similarity
Belongs to the calreticulin family.
Uniprot
H9IV17
A0A2A4K368
A0A3S2M6S4
A0A2H1WSR5
A0A212ETQ6
S4PJP8
+ More
A0A194Q687 A0A1B6DDQ8 A0A1B6EDM4 A0A1B6JET0 A0A0T6B1K4 A0A067QU32 A0A2J7RG58 D1ZZU8 A0A0P5XWM1 A0A0N8CQX6 A0A0P6FVL7 A0A1Y1K457 A0A164QKC8 A0A182PRF1 A0A0N8B5C7 E9GMP2 A0A0N8D563 A0A0P5HRB7 Q7Q9V3 A0A0P5MEV0 A0A0P5NZQ1 A0A0N8CD09 A0A0N8EFW9 A0A0P5JRX1 A0A182RUV5 A0A0N8DYE7 A0A0P5VA39 A0A0P5S809 A0A0P4YJW6 A0A0N8CHT9 A0A0P5W9K8 A0A0P5NC16 A0A0P5P8F8 B0WPL8 A0A0P5P1L5 A0A0P5SIY8 A0A0P5GY15 A0A0P4Z8P3 Q1DH50 A0A0P4YQG7 A0A0N7ZVC6 A0A0P5JU82 A0A0P4XYX2 A0A1W7R7X3 A0A1S4G3R0 A0A0P5SH25 A0A1B0CQP7 A0A0P5XP00 A0A0P5ZRQ7 A0A0N8C765 U5EYL6 A0A0P5NX68 V5I8J4 E0W030 A0A182JBP8 A0A2M4A828 A0A182WM89 A0A2M4A8B3 A0A2M4A897 A0A2M4A824 A0A1Q3FIK1 A0A1Q3FIH6 W5JI33 A0A2M4BHQ3 A0A0P5YPT0 A0A2M4BHS0 A0A2M4CMF7 A0A1L8DU76 A0A2M3ZE08 A0A084WDP7 A0A1L8DU51 A0A193DUV6 A0A023EUG1 A0A232EMT2 A0A034W3J9 A0A2M3Z340 A0A0K8U2X7 A0A0K8UZT9 T1PBX2 A0A075LY63 A0A182LB29 A0A1I8M1P1 A0A0B4J2Q3 U4TVI5 A0A0P4YMS0 A0A0P4XK38 A0A034W0Z1 A0A0A1XN99 A0A0N8BPS4 A0A182HPG0 A0A0P5ZWW2 A0A182JXZ6 A0A0P5XNZ3 A7UT70
A0A194Q687 A0A1B6DDQ8 A0A1B6EDM4 A0A1B6JET0 A0A0T6B1K4 A0A067QU32 A0A2J7RG58 D1ZZU8 A0A0P5XWM1 A0A0N8CQX6 A0A0P6FVL7 A0A1Y1K457 A0A164QKC8 A0A182PRF1 A0A0N8B5C7 E9GMP2 A0A0N8D563 A0A0P5HRB7 Q7Q9V3 A0A0P5MEV0 A0A0P5NZQ1 A0A0N8CD09 A0A0N8EFW9 A0A0P5JRX1 A0A182RUV5 A0A0N8DYE7 A0A0P5VA39 A0A0P5S809 A0A0P4YJW6 A0A0N8CHT9 A0A0P5W9K8 A0A0P5NC16 A0A0P5P8F8 B0WPL8 A0A0P5P1L5 A0A0P5SIY8 A0A0P5GY15 A0A0P4Z8P3 Q1DH50 A0A0P4YQG7 A0A0N7ZVC6 A0A0P5JU82 A0A0P4XYX2 A0A1W7R7X3 A0A1S4G3R0 A0A0P5SH25 A0A1B0CQP7 A0A0P5XP00 A0A0P5ZRQ7 A0A0N8C765 U5EYL6 A0A0P5NX68 V5I8J4 E0W030 A0A182JBP8 A0A2M4A828 A0A182WM89 A0A2M4A8B3 A0A2M4A897 A0A2M4A824 A0A1Q3FIK1 A0A1Q3FIH6 W5JI33 A0A2M4BHQ3 A0A0P5YPT0 A0A2M4BHS0 A0A2M4CMF7 A0A1L8DU76 A0A2M3ZE08 A0A084WDP7 A0A1L8DU51 A0A193DUV6 A0A023EUG1 A0A232EMT2 A0A034W3J9 A0A2M3Z340 A0A0K8U2X7 A0A0K8UZT9 T1PBX2 A0A075LY63 A0A182LB29 A0A1I8M1P1 A0A0B4J2Q3 U4TVI5 A0A0P4YMS0 A0A0P4XK38 A0A034W0Z1 A0A0A1XN99 A0A0N8BPS4 A0A182HPG0 A0A0P5ZWW2 A0A182JXZ6 A0A0P5XNZ3 A7UT70
Pubmed
EMBL
BABH01000158
NWSH01000210
PCG78368.1
RSAL01000023
RVE52256.1
ODYU01010728
+ More
SOQ56016.1 AGBW02012542 OWR44859.1 GAIX01004860 JAA87700.1 KQ459439 KPJ01047.1 GEDC01013512 JAS23786.1 GEDC01001249 JAS36049.1 GECU01009992 JAS97714.1 LJIG01016317 KRT80965.1 KK853449 KDR07563.1 NEVH01004406 PNF39813.1 KQ971338 EFA01796.1 GDIP01066252 JAM37463.1 GDIP01107639 JAL96075.1 GDIQ01042165 JAN52572.1 GEZM01095908 GEZM01095907 GEZM01095906 JAV55271.1 LRGB01002384 KZS07835.1 GDIQ01211243 JAK40482.1 GL732553 EFX79258.1 GDIP01067743 JAM35972.1 GDIQ01246983 JAK04742.1 AAAB01008900 EAA09483.4 GDIQ01160562 JAK91163.1 GDIQ01145572 JAL06154.1 GDIQ01192199 GDIQ01091787 JAL59939.1 GDIQ01030716 JAN64021.1 GDIP01158830 GDIQ01195896 GDIP01122035 GDIP01120237 GDIQ01052470 GDIQ01044468 JAJ64572.1 JAK55829.1 GDIQ01079692 JAN15045.1 GDIP01104988 JAL98726.1 GDIQ01108140 JAL43586.1 GDIP01228672 JAI94729.1 GDIP01130335 JAL73379.1 GDIP01089552 JAM14163.1 GDIQ01144229 JAL07497.1 GDIQ01136471 JAL15255.1 DS232027 EDS32421.1 GDIQ01136472 JAL15254.1 GDIP01139238 JAL64476.1 GDIQ01240948 JAK10777.1 GDIP01217877 JAJ05525.1 CH899801 EAT32491.1 GDIP01224026 JAI99375.1 GDIP01208929 JAJ14473.1 GDIQ01193952 JAK57773.1 GDIP01236983 JAI86418.1 GEHC01000383 JAV47262.1 GDIQ01091788 JAL59938.1 AJWK01023835 AJWK01023836 GDIP01069341 JAM34374.1 GDIP01040258 JAM63457.1 GDIQ01108139 JAL43587.1 GANO01000276 JAB59595.1 GDIQ01136470 JAL15256.1 GALX01004580 JAB63886.1 DS235855 EEB18986.1 GGFK01003633 MBW36954.1 GGFK01003684 MBW37005.1 GGFK01003703 MBW37024.1 GGFK01003632 MBW36953.1 GFDL01007689 JAV27356.1 GFDL01007687 JAV27358.1 ADMH02001185 ETN63791.1 GGFJ01003445 MBW52586.1 GDIP01056147 JAM47568.1 GGFJ01003446 MBW52587.1 GGFL01002339 MBW66517.1 GFDF01004114 JAV09970.1 GGFM01005967 MBW26718.1 ATLV01023051 KE525340 KFB48341.1 GFDF01004115 JAV09969.1 KU519399 ANN46488.1 GAPW01001032 JAC12566.1 NNAY01003301 OXU19656.1 GAKP01010604 JAC48348.1 GGFM01002164 MBW22915.1 GDHF01031436 JAI20878.1 GDHF01020241 GDHF01018072 JAI32073.1 JAI34242.1 KA646266 AFP60895.1 KJ914578 AIF71174.1 KB631724 ERL85594.1 GDIP01226475 JAI96926.1 GDIP01240786 GDIP01122034 GDIQ01052469 GDIQ01049560 JAI82615.1 JAN42268.1 GAKP01010603 JAC48349.1 GBXI01001860 JAD12432.1 GDIQ01156867 JAK94858.1 APCN01002995 GDIP01050852 JAM52863.1 GDIP01069342 JAM34373.1 EDO64053.1
SOQ56016.1 AGBW02012542 OWR44859.1 GAIX01004860 JAA87700.1 KQ459439 KPJ01047.1 GEDC01013512 JAS23786.1 GEDC01001249 JAS36049.1 GECU01009992 JAS97714.1 LJIG01016317 KRT80965.1 KK853449 KDR07563.1 NEVH01004406 PNF39813.1 KQ971338 EFA01796.1 GDIP01066252 JAM37463.1 GDIP01107639 JAL96075.1 GDIQ01042165 JAN52572.1 GEZM01095908 GEZM01095907 GEZM01095906 JAV55271.1 LRGB01002384 KZS07835.1 GDIQ01211243 JAK40482.1 GL732553 EFX79258.1 GDIP01067743 JAM35972.1 GDIQ01246983 JAK04742.1 AAAB01008900 EAA09483.4 GDIQ01160562 JAK91163.1 GDIQ01145572 JAL06154.1 GDIQ01192199 GDIQ01091787 JAL59939.1 GDIQ01030716 JAN64021.1 GDIP01158830 GDIQ01195896 GDIP01122035 GDIP01120237 GDIQ01052470 GDIQ01044468 JAJ64572.1 JAK55829.1 GDIQ01079692 JAN15045.1 GDIP01104988 JAL98726.1 GDIQ01108140 JAL43586.1 GDIP01228672 JAI94729.1 GDIP01130335 JAL73379.1 GDIP01089552 JAM14163.1 GDIQ01144229 JAL07497.1 GDIQ01136471 JAL15255.1 DS232027 EDS32421.1 GDIQ01136472 JAL15254.1 GDIP01139238 JAL64476.1 GDIQ01240948 JAK10777.1 GDIP01217877 JAJ05525.1 CH899801 EAT32491.1 GDIP01224026 JAI99375.1 GDIP01208929 JAJ14473.1 GDIQ01193952 JAK57773.1 GDIP01236983 JAI86418.1 GEHC01000383 JAV47262.1 GDIQ01091788 JAL59938.1 AJWK01023835 AJWK01023836 GDIP01069341 JAM34374.1 GDIP01040258 JAM63457.1 GDIQ01108139 JAL43587.1 GANO01000276 JAB59595.1 GDIQ01136470 JAL15256.1 GALX01004580 JAB63886.1 DS235855 EEB18986.1 GGFK01003633 MBW36954.1 GGFK01003684 MBW37005.1 GGFK01003703 MBW37024.1 GGFK01003632 MBW36953.1 GFDL01007689 JAV27356.1 GFDL01007687 JAV27358.1 ADMH02001185 ETN63791.1 GGFJ01003445 MBW52586.1 GDIP01056147 JAM47568.1 GGFJ01003446 MBW52587.1 GGFL01002339 MBW66517.1 GFDF01004114 JAV09970.1 GGFM01005967 MBW26718.1 ATLV01023051 KE525340 KFB48341.1 GFDF01004115 JAV09969.1 KU519399 ANN46488.1 GAPW01001032 JAC12566.1 NNAY01003301 OXU19656.1 GAKP01010604 JAC48348.1 GGFM01002164 MBW22915.1 GDHF01031436 JAI20878.1 GDHF01020241 GDHF01018072 JAI32073.1 JAI34242.1 KA646266 AFP60895.1 KJ914578 AIF71174.1 KB631724 ERL85594.1 GDIP01226475 JAI96926.1 GDIP01240786 GDIP01122034 GDIQ01052469 GDIQ01049560 JAI82615.1 JAN42268.1 GAKP01010603 JAC48349.1 GBXI01001860 JAD12432.1 GDIQ01156867 JAK94858.1 APCN01002995 GDIP01050852 JAM52863.1 GDIP01069342 JAM34373.1 EDO64053.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000027135
+ More
UP000235965 UP000007266 UP000076858 UP000075885 UP000000305 UP000007062 UP000075900 UP000002320 UP000008820 UP000092461 UP000009046 UP000075880 UP000075920 UP000000673 UP000030765 UP000215335 UP000075882 UP000095301 UP000002358 UP000030742 UP000075840 UP000075881
UP000235965 UP000007266 UP000076858 UP000075885 UP000000305 UP000007062 UP000075900 UP000002320 UP000008820 UP000092461 UP000009046 UP000075880 UP000075920 UP000000673 UP000030765 UP000215335 UP000075882 UP000095301 UP000002358 UP000030742 UP000075840 UP000075881
PRIDE
Pfam
PF00262 Calreticulin
Interpro
Gene 3D
ProteinModelPortal
H9IV17
A0A2A4K368
A0A3S2M6S4
A0A2H1WSR5
A0A212ETQ6
S4PJP8
+ More
A0A194Q687 A0A1B6DDQ8 A0A1B6EDM4 A0A1B6JET0 A0A0T6B1K4 A0A067QU32 A0A2J7RG58 D1ZZU8 A0A0P5XWM1 A0A0N8CQX6 A0A0P6FVL7 A0A1Y1K457 A0A164QKC8 A0A182PRF1 A0A0N8B5C7 E9GMP2 A0A0N8D563 A0A0P5HRB7 Q7Q9V3 A0A0P5MEV0 A0A0P5NZQ1 A0A0N8CD09 A0A0N8EFW9 A0A0P5JRX1 A0A182RUV5 A0A0N8DYE7 A0A0P5VA39 A0A0P5S809 A0A0P4YJW6 A0A0N8CHT9 A0A0P5W9K8 A0A0P5NC16 A0A0P5P8F8 B0WPL8 A0A0P5P1L5 A0A0P5SIY8 A0A0P5GY15 A0A0P4Z8P3 Q1DH50 A0A0P4YQG7 A0A0N7ZVC6 A0A0P5JU82 A0A0P4XYX2 A0A1W7R7X3 A0A1S4G3R0 A0A0P5SH25 A0A1B0CQP7 A0A0P5XP00 A0A0P5ZRQ7 A0A0N8C765 U5EYL6 A0A0P5NX68 V5I8J4 E0W030 A0A182JBP8 A0A2M4A828 A0A182WM89 A0A2M4A8B3 A0A2M4A897 A0A2M4A824 A0A1Q3FIK1 A0A1Q3FIH6 W5JI33 A0A2M4BHQ3 A0A0P5YPT0 A0A2M4BHS0 A0A2M4CMF7 A0A1L8DU76 A0A2M3ZE08 A0A084WDP7 A0A1L8DU51 A0A193DUV6 A0A023EUG1 A0A232EMT2 A0A034W3J9 A0A2M3Z340 A0A0K8U2X7 A0A0K8UZT9 T1PBX2 A0A075LY63 A0A182LB29 A0A1I8M1P1 A0A0B4J2Q3 U4TVI5 A0A0P4YMS0 A0A0P4XK38 A0A034W0Z1 A0A0A1XN99 A0A0N8BPS4 A0A182HPG0 A0A0P5ZWW2 A0A182JXZ6 A0A0P5XNZ3 A7UT70
A0A194Q687 A0A1B6DDQ8 A0A1B6EDM4 A0A1B6JET0 A0A0T6B1K4 A0A067QU32 A0A2J7RG58 D1ZZU8 A0A0P5XWM1 A0A0N8CQX6 A0A0P6FVL7 A0A1Y1K457 A0A164QKC8 A0A182PRF1 A0A0N8B5C7 E9GMP2 A0A0N8D563 A0A0P5HRB7 Q7Q9V3 A0A0P5MEV0 A0A0P5NZQ1 A0A0N8CD09 A0A0N8EFW9 A0A0P5JRX1 A0A182RUV5 A0A0N8DYE7 A0A0P5VA39 A0A0P5S809 A0A0P4YJW6 A0A0N8CHT9 A0A0P5W9K8 A0A0P5NC16 A0A0P5P8F8 B0WPL8 A0A0P5P1L5 A0A0P5SIY8 A0A0P5GY15 A0A0P4Z8P3 Q1DH50 A0A0P4YQG7 A0A0N7ZVC6 A0A0P5JU82 A0A0P4XYX2 A0A1W7R7X3 A0A1S4G3R0 A0A0P5SH25 A0A1B0CQP7 A0A0P5XP00 A0A0P5ZRQ7 A0A0N8C765 U5EYL6 A0A0P5NX68 V5I8J4 E0W030 A0A182JBP8 A0A2M4A828 A0A182WM89 A0A2M4A8B3 A0A2M4A897 A0A2M4A824 A0A1Q3FIK1 A0A1Q3FIH6 W5JI33 A0A2M4BHQ3 A0A0P5YPT0 A0A2M4BHS0 A0A2M4CMF7 A0A1L8DU76 A0A2M3ZE08 A0A084WDP7 A0A1L8DU51 A0A193DUV6 A0A023EUG1 A0A232EMT2 A0A034W3J9 A0A2M3Z340 A0A0K8U2X7 A0A0K8UZT9 T1PBX2 A0A075LY63 A0A182LB29 A0A1I8M1P1 A0A0B4J2Q3 U4TVI5 A0A0P4YMS0 A0A0P4XK38 A0A034W0Z1 A0A0A1XN99 A0A0N8BPS4 A0A182HPG0 A0A0P5ZWW2 A0A182JXZ6 A0A0P5XNZ3 A7UT70
PDB
1JHN
E-value=1.88637e-125,
Score=1151
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum
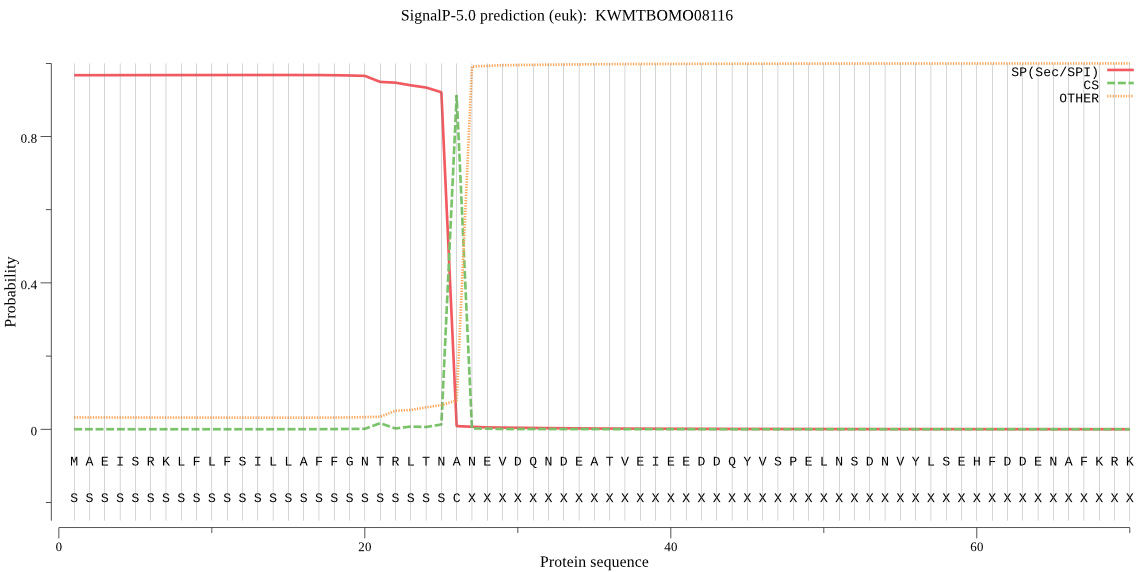
SignalP
Position: 1 - 26,
Likelihood: 0.968124
Length:
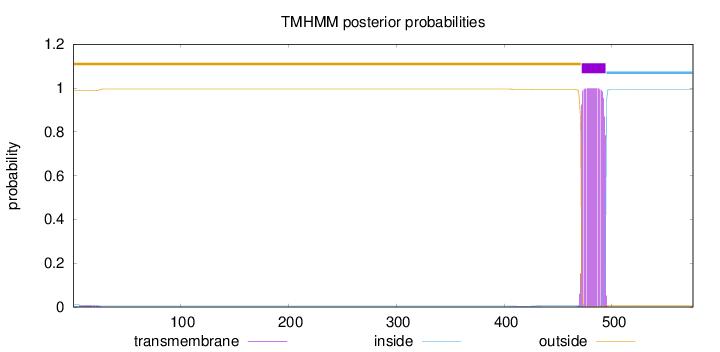
575
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.92669
Exp number, first 60 AAs:
0.13153
Total prob of N-in:
0.01143
outside
1 - 471
TMhelix
472 - 494
inside
495 - 575
Population Genetic Test Statistics
Pi
21.543334
Theta
15.578143
Tajima's D
-1.301776
CLR
13.978768
CSRT
0.0825458727063647
Interpretation
Uncertain
