Pre Gene Modal
BGIBMGA001100
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_tyrosine-protein_phosphatase_non-receptor_type_11-like_[Bombyx_mori]
Full name
Tyrosine-protein phosphatase non-receptor type
Location in the cell
Cytoplasmic Reliability : 2.757
Sequence
CDS
ATGATAACTCGAAGATGGTTCCACCCCTCGCTGAATGGTGTGGACGCGGAAAAATTGCTCATGGAGTGCGGTCACGATGGGTATTTCCTGGCCAGGCCAAGTTCAAGTAATAAAGGAGATTTCACTCTCTCCGTCCGAAGAGGAAATGAGGTCACTCATATAAAAATTCAGAACAACGGCGAATTTCTCGATCTTTATGGAGGTGAAAAATTTGCTACCCTGTCAGAACTCGTCCAATATTATATGGACAATCAATGCCAGCTAAGAGAAAAAAATGGAAATATAATTAGGTTAAAAACGCCTCTCAACTGTGCAGATCCAACAACCGAACGATGGTACCACGGGCAGCTCACTGCCAAGGAAGCAGAGAGAATGATGATGGAAAACGGAAAAAACGGGTCGTTTCTCGTTCGCGAATCCCAAAGGCAACCTGGCGACTTCGTGTTGTCCGTTCGCACGAGAGATCGAGTTACACATGTGATAATTAGGCGAAAGGACAACAAATATGACGTAGGTGGGGGGCAACAGTTTGACGATCTCGTAAGTCTAATAGAATACTATCGTAGTTTTCCCATGGTCGAGACTACTGGTGAGGAAAACGAGGGACCCATCGAGAGCATGGCCTACAAACAAGGTTTCTGGGAGGAATTCGAAACGTTACAGATGATGGAGAACTTGCAGTTGTTCGACAGAATGGAGGGTTCGAAACCGGAGAATATTAGGAAAAACAGATACAAAAATATTATACCATTCGATCACACGCGTGTTATTTTAAAGGACATCCCCCCTGACGGTCCTCCGGGCTCAGATTACATCAATGCCAACTACATTCGATGTGACAGTATGGATTCCATTTCCGATTCCCAAGAATTCACAGGGAACGGCAGCACTGAAAACGGCAAAGACGGTACGCCGTCGAAAGCTAAAGATAAATCATCGCCTGTTCATACAAGCGTTATCGTAACTGAAGAGCCGGTCAAATCAAGTAAGAAGGTGCACGGCAACGGAACACACAAGCTGCCGGCGTTCGAACCGAGCGTTCTTCGTCCCAACCCGAATTATTTCAATACGACAATACCGAAATCAGCGACCGAAACCGAAAACGGTGTGCCCACTGTACACGTGTACAACAAAACGTACATAGCTACTCAAGGCTGTCTTTCGACAACAATATACCCGTTCTGGTCGATGATATGGCAAGAGGACGTCCGCATCATCATAATGACGACAAAGGAGATCGAGCGCGGGAAGGTCAAATGCGAAAGGTACTGGCCGGACTTGAACAAAACAGAGGTCGTCAAGAAGTACACGATACTTAACGAATTCGAATCCTCCACGCCGGATTATACGCTTCGACGGTTCCTTGTCACGAAAAAAGATGAGACTACAGTGAAGCGGACCATATATCATTTCCATTTCACGGCCTGGCCGGACCATCGTGTTCCCTCAGAGCCCGGTCGCGTGCTCAACATTCTACTGGACGTGAACTACAGGCTACAGCAGATAATGACGGGGACCGATCCTCCGGCGCAGGCGGTCGTATGTGTTCATTGCTCGGCGGGTATAGGTCGGACGGGAACCTTCATAGTCATCGATATGATCTTGGACCAAATAAGAAAGGAAGGGTTCGACTGCGAGATCGACATACATCGCACTGTGCAGATGGTGCGCGACCAAAGGTCCGGTATGGTGCAGAACGAAGCTCAATATAAGTTTATCTACATGGCCGTGTTGGAGTTTATCGAAACGGAGAAGCAAAGGGTAGGTCTCGGGCCGGAGGCGGCACAGGACTCGCCGCGAGCCAGGTCTATGCCCGTACCATTTTTATGA
Protein
MITRRWFHPSLNGVDAEKLLMECGHDGYFLARPSSSNKGDFTLSVRRGNEVTHIKIQNNGEFLDLYGGEKFATLSELVQYYMDNQCQLREKNGNIIRLKTPLNCADPTTERWYHGQLTAKEAERMMMENGKNGSFLVRESQRQPGDFVLSVRTRDRVTHVIIRRKDNKYDVGGGQQFDDLVSLIEYYRSFPMVETTGEENEGPIESMAYKQGFWEEFETLQMMENLQLFDRMEGSKPENIRKNRYKNIIPFDHTRVILKDIPPDGPPGSDYINANYIRCDSMDSISDSQEFTGNGSTENGKDGTPSKAKDKSSPVHTSVIVTEEPVKSSKKVHGNGTHKLPAFEPSVLRPNPNYFNTTIPKSATETENGVPTVHVYNKTYIATQGCLSTTIYPFWSMIWQEDVRIIIMTTKEIERGKVKCERYWPDLNKTEVVKKYTILNEFESSTPDYTLRRFLVTKKDETTVKRTIYHFHFTAWPDHRVPSEPGRVLNILLDVNYRLQQIMTGTDPPAQAVVCVHCSAGIGRTGTFIVIDMILDQIRKEGFDCEIDIHRTVQMVRDQRSGMVQNEAQYKFIYMAVLEFIETEKQRVGLGPEAAQDSPRARSMPVPFL
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class 2 subfamily.
Uniprot
H9IV20
A0A2H1WY52
A0A2A4K380
A0A194Q681
A0A194RJU9
A0A0L7LLZ9
+ More
A0A212ETM7 A0A1Q3FV42 A0A1Q3FUP0 A0A1Q3FV66 A0A1S4FJQ9 A0A1L8DLN1 A0A1J1HPS2 A0A1J1HV72 A0A195DM14 A0A1I8MWH3 A0A1S4GFJ7 A0A0K8UG39 A0A0A1XPL0 A0A034V7D6 A0A034V5R1 W8BRK0 W8BL49 W8BDZ7 Q5TTR3 A0A2M3ZZK1 A0A2M4BH42 A0A0A1WVB4 A0A0P6DZ20 A0A0R3NZB4 A0A0R3NZM3 A0A452C922 A0A2Y9QLI1 A0A2U4CKF1 A0A452C8W1 A0A2Y9QEG1 A0A2U4CKM6 A0A0V1HZH6 A0A0V1DZC7 A0A1Q3FXY5 A0A1Q3FY24 A0A1J1HR93 A0A1L8DLU8 A0A1Q3FVE6 A0A1Q3FV89 A0A1A9W0Q5 A0A3S2M4Z3 A0A0K8VQ68 A0A0K8VYW7 A0A2A4JVZ2 A0A2A4JVA9 A0A1Q3FY41 A0A1A9YBK0 A0A1B0D637
A0A212ETM7 A0A1Q3FV42 A0A1Q3FUP0 A0A1Q3FV66 A0A1S4FJQ9 A0A1L8DLN1 A0A1J1HPS2 A0A1J1HV72 A0A195DM14 A0A1I8MWH3 A0A1S4GFJ7 A0A0K8UG39 A0A0A1XPL0 A0A034V7D6 A0A034V5R1 W8BRK0 W8BL49 W8BDZ7 Q5TTR3 A0A2M3ZZK1 A0A2M4BH42 A0A0A1WVB4 A0A0P6DZ20 A0A0R3NZB4 A0A0R3NZM3 A0A452C922 A0A2Y9QLI1 A0A2U4CKF1 A0A452C8W1 A0A2Y9QEG1 A0A2U4CKM6 A0A0V1HZH6 A0A0V1DZC7 A0A1Q3FXY5 A0A1Q3FY24 A0A1J1HR93 A0A1L8DLU8 A0A1Q3FVE6 A0A1Q3FV89 A0A1A9W0Q5 A0A3S2M4Z3 A0A0K8VQ68 A0A0K8VYW7 A0A2A4JVZ2 A0A2A4JVA9 A0A1Q3FY41 A0A1A9YBK0 A0A1B0D637
EC Number
3.1.3.48
Pubmed
EMBL
BABH01000164
ODYU01011528
SOQ57314.1
NWSH01000210
PCG78378.1
KQ459439
+ More
KPJ01043.1 KQ460297 KPJ16226.1 JTDY01000610 KOB76472.1 AGBW02012542 OWR44855.1 GFDL01003723 JAV31322.1 GFDL01003724 JAV31321.1 GFDL01003722 JAV31323.1 GFDF01006807 JAV07277.1 CVRI01000015 CRK90056.1 CRK90057.1 KQ980762 KYN13534.1 AAAB01008859 GDHF01027049 JAI25265.1 GBXI01001512 JAD12780.1 GAKP01021479 JAC37473.1 GAKP01021475 JAC37477.1 GAMC01007072 JAB99483.1 GAMC01007073 JAB99482.1 GAMC01007070 JAB99485.1 EAL40926.4 GGFK01000610 MBW33931.1 GGFJ01003172 MBW52313.1 GBXI01011711 JAD02581.1 GDIQ01070546 JAN24191.1 CH379064 KRT06376.1 KRT06377.1 JYDS01000310 KRZ15474.1 JYDR01000169 KRY66434.1 GFDL01002679 JAV32366.1 GFDL01002632 JAV32413.1 CRK90058.1 GFDF01006750 JAV07334.1 GFDL01003527 JAV31518.1 GFDL01003526 JAV31519.1 RSAL01000032 RVE51530.1 GDHF01011326 JAI40988.1 GDHF01008247 JAI44067.1 NWSH01000575 PCG75552.1 PCG75554.1 GFDL01002643 JAV32402.1 AJVK01025699 AJVK01025700 AJVK01025701 AJVK01025702
KPJ01043.1 KQ460297 KPJ16226.1 JTDY01000610 KOB76472.1 AGBW02012542 OWR44855.1 GFDL01003723 JAV31322.1 GFDL01003724 JAV31321.1 GFDL01003722 JAV31323.1 GFDF01006807 JAV07277.1 CVRI01000015 CRK90056.1 CRK90057.1 KQ980762 KYN13534.1 AAAB01008859 GDHF01027049 JAI25265.1 GBXI01001512 JAD12780.1 GAKP01021479 JAC37473.1 GAKP01021475 JAC37477.1 GAMC01007072 JAB99483.1 GAMC01007073 JAB99482.1 GAMC01007070 JAB99485.1 EAL40926.4 GGFK01000610 MBW33931.1 GGFJ01003172 MBW52313.1 GBXI01011711 JAD02581.1 GDIQ01070546 JAN24191.1 CH379064 KRT06376.1 KRT06377.1 JYDS01000310 KRZ15474.1 JYDR01000169 KRY66434.1 GFDL01002679 JAV32366.1 GFDL01002632 JAV32413.1 CRK90058.1 GFDF01006750 JAV07334.1 GFDL01003527 JAV31518.1 GFDL01003526 JAV31519.1 RSAL01000032 RVE51530.1 GDHF01011326 JAI40988.1 GDHF01008247 JAI44067.1 NWSH01000575 PCG75552.1 PCG75554.1 GFDL01002643 JAV32402.1 AJVK01025699 AJVK01025700 AJVK01025701 AJVK01025702
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IV20
A0A2H1WY52
A0A2A4K380
A0A194Q681
A0A194RJU9
A0A0L7LLZ9
+ More
A0A212ETM7 A0A1Q3FV42 A0A1Q3FUP0 A0A1Q3FV66 A0A1S4FJQ9 A0A1L8DLN1 A0A1J1HPS2 A0A1J1HV72 A0A195DM14 A0A1I8MWH3 A0A1S4GFJ7 A0A0K8UG39 A0A0A1XPL0 A0A034V7D6 A0A034V5R1 W8BRK0 W8BL49 W8BDZ7 Q5TTR3 A0A2M3ZZK1 A0A2M4BH42 A0A0A1WVB4 A0A0P6DZ20 A0A0R3NZB4 A0A0R3NZM3 A0A452C922 A0A2Y9QLI1 A0A2U4CKF1 A0A452C8W1 A0A2Y9QEG1 A0A2U4CKM6 A0A0V1HZH6 A0A0V1DZC7 A0A1Q3FXY5 A0A1Q3FY24 A0A1J1HR93 A0A1L8DLU8 A0A1Q3FVE6 A0A1Q3FV89 A0A1A9W0Q5 A0A3S2M4Z3 A0A0K8VQ68 A0A0K8VYW7 A0A2A4JVZ2 A0A2A4JVA9 A0A1Q3FY41 A0A1A9YBK0 A0A1B0D637
A0A212ETM7 A0A1Q3FV42 A0A1Q3FUP0 A0A1Q3FV66 A0A1S4FJQ9 A0A1L8DLN1 A0A1J1HPS2 A0A1J1HV72 A0A195DM14 A0A1I8MWH3 A0A1S4GFJ7 A0A0K8UG39 A0A0A1XPL0 A0A034V7D6 A0A034V5R1 W8BRK0 W8BL49 W8BDZ7 Q5TTR3 A0A2M3ZZK1 A0A2M4BH42 A0A0A1WVB4 A0A0P6DZ20 A0A0R3NZB4 A0A0R3NZM3 A0A452C922 A0A2Y9QLI1 A0A2U4CKF1 A0A452C8W1 A0A2Y9QEG1 A0A2U4CKM6 A0A0V1HZH6 A0A0V1DZC7 A0A1Q3FXY5 A0A1Q3FY24 A0A1J1HR93 A0A1L8DLU8 A0A1Q3FVE6 A0A1Q3FV89 A0A1A9W0Q5 A0A3S2M4Z3 A0A0K8VQ68 A0A0K8VYW7 A0A2A4JVZ2 A0A2A4JVA9 A0A1Q3FY41 A0A1A9YBK0 A0A1B0D637
PDB
6MDD
E-value=1.56351e-97,
Score=911
Ontologies
GO
Topology
Subcellular location
Cytoplasm
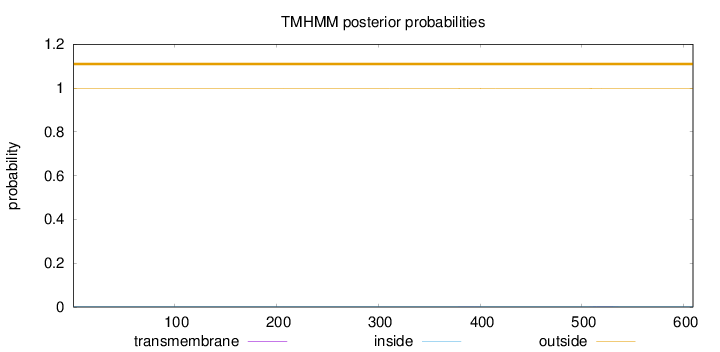
Length:
609
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05924
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00114
outside
1 - 609
Population Genetic Test Statistics
Pi
21.987536
Theta
4.482022
Tajima's D
0.502643
CLR
1.500357
CSRT
0.543922803859807
Interpretation
Uncertain
