Gene
KWMTBOMO08109
Pre Gene Modal
BGIBMGA001101
Annotation
PREDICTED:_TBC1_domain_family_member_19_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.195
Sequence
CDS
ATGGACGGGGTTAAAGAAGAAAAGCTAGTAGCATCGCCAAATGTGAAAAAAGAAGACTTTAAACAAACGCTGCAGCAGGCTATGAAAGAAAAGGGATTAGACACGAAATTAAGGAACACTGTTTTCCATTGGGTTCGAACGCAAAGTAAACAGAATAAATTAGATCCTTTAACTTCACTATCAAAAGCGAGCGCACAATGGGAGAAGAGAATACACAAATCGTTAAATTCCATGTGTTCAGATCTCGAAACGTCATTAGCGAAAATACGGCCACAAAGCGAACAGGACGAATTAGCTGATAAATGGAATGAATTGAGCACATACAATTTAGATTTAACAAAGTACCGGCCAGTTTACGCTCCGAAAGATTTTTTAGAAGTTTTGTTAACACTATCCGGATATGTTCCATTCACGAGAGAAGACGAGCCAAAGTGGGAATTCGCGCATCTACCCCTTCAGGTGAAGACTTTAGATCAAATCAGAAAATTATACTTGGAGTGGTCAAGCGGTGAGCCGCTGTTAGGCGCTAACCCGCACATGGCGAGCGCAGTGCCAGGGTTCGTCACTCTAGAGGCCGAGCGTGTCAGCTTGGGCGAAAGAGTGGCGGCTCTGGGCTACGCACCCGTGGTACAGGAATATTTGAAGAAGGGCAGCCCACAGTGTCTAAGGGCCAAATTGTGGGCTCAAGTGCTAGGCTCGGAGATCAAGGGACAGCAAATAACATATTTCAATCAACTAAAGAAAAGTGTTCTGGAAGTGGATCTGATGATAGACAAGTTAATATTCAAAGACGTGCAGTTAACCGCCTCGAACGATGATCAATATTTTGTATTCGAAGATGTATTATACCAGGTAATGTTATGTTTCTCCCGAGACTGCGACGTGATGCAGCAGCTCAAGGGCAGCATCGGTAGTCCTCTCACGGTCACTATAAAGGGCAAACAGACATCCACCGACAGCCTCACAGTCTTCCCTCCCAGCGGGGTCATACCTTTTCACGGCTTTACTATGTATGCAACTCCCTTTTGTTATTTGTACGACGATCCCGTGCAACTTTACTACACGTTTCGAGCGTTCTACATAAGGTACTGGCATCGACTACATTACATATCGACACATCCACAGGGTATAGTATCGTTATGCCTTTTGTACGAGAGGCTTTTGGAAGCGAACGAACCTTTGTTATGGATACATTTCAGAAATATAAACATTAACCCAATCAGAGTTGTATTCAAGTGGTTAATGCGGGCATTCAGTGGGCACCTACCGCCGGATCAACTGTTACTATTATGGGATGCGATACTAGGTTACGACTGCCTAGAAATTCTGCCGTTGCTGGCGCTGGCGATATTGAGTTTCAGGAAAGAGAATATCTTCCAAGTTAATACGTTGCAGAATGTCGACGCTGTTTTAGCCGATCTATCGACTATATCGGTTATACCGCTTCTGCAATTGGCTTTGATGAGAGCTTAG
Protein
MDGVKEEKLVASPNVKKEDFKQTLQQAMKEKGLDTKLRNTVFHWVRTQSKQNKLDPLTSLSKASAQWEKRIHKSLNSMCSDLETSLAKIRPQSEQDELADKWNELSTYNLDLTKYRPVYAPKDFLEVLLTLSGYVPFTREDEPKWEFAHLPLQVKTLDQIRKLYLEWSSGEPLLGANPHMASAVPGFVTLEAERVSLGERVAALGYAPVVQEYLKKGSPQCLRAKLWAQVLGSEIKGQQITYFNQLKKSVLEVDLMIDKLIFKDVQLTASNDDQYFVFEDVLYQVMLCFSRDCDVMQQLKGSIGSPLTVTIKGKQTSTDSLTVFPPSGVIPFHGFTMYATPFCYLYDDPVQLYYTFRAFYIRYWHRLHYISTHPQGIVSLCLLYERLLEANEPLLWIHFRNININPIRVVFKWLMRAFSGHLPPDQLLLLWDAILGYDCLEILPLLALAILSFRKENIFQVNTLQNVDAVLADLSTISVIPLLQLALMRA
Summary
Uniprot
A0A2H1WWF4
A0A437BPH5
A0A2A4K393
A0A212ETN2
A0A194QC78
A0A1B6GZ25
+ More
A0A1B6KZC5 A0A1B6HSG2 A0A194RFI9 A0A1B6D6P0 A0A088A4M7 D2A1I5 A0A1B6GGU9 A0A1B6JD21 A0A0C9PVE1 A0A2P8ZCY9 A0A1B6I016 K7ISA4 A0A195E5N0 E2BLK2 A0A232ENH6 A0A0N0U3F4 A0A151HZ23 A0A0L7R3A0 E2AJP4 F4W4C9 A0A151WXE6 A0A1B6II49 A0A026WJ32 A0A2A3E9C5 T1HK41 U4UMS3 A0A154PQI7 A0A151ILV7 A0A1S4E7E1 U4UN97 A0A067QYJ8 A0A2J7RRW7 A0A1B6M8T7 J9JPK9 E0W319 A0A1I8NVI9 A0A1I8NVJ3 A0A336LY43 A0A0K2T1U8 A0A2S2NAL0 A0A1A9VMJ0 A0A0L0CDH8 B4JDH8 A0A182RDQ9 B4P085 A0A182JF88 B3N4T7 B4N788 A0A182W6G1 A0A182LXR0 A0A310SNH7 A0A1I8NVI7 A0A034W9X2 Q9VMT4 B4Q3J6 A0A0K8TZJ8 A0A182YDU8 A0A084WH79 B4LSN2 A0A1B0AEK7 A0A182L0J9 B4I1E1 A0A182IC70 Q7Q4F7 A0A182XFY3 A0A1W4UV89 Q16YB1 A0A182PLR0 A0A182TRN9 A0A3B0JB05 A0A182NTZ3 A0A182JT46 W8B575 A0A182Q904 B3MLM7 A0A182V911 A0A1S4GYS3 W5JQK2 B4G6P9 Q29M19 J9JU69 B4KG17 A0A0M4E5E9 A0A1A9Y3H1 A0A1Q3G5C7 A0A182G4P9 A0A1J1IJZ5 A0A182F3F6 A0A1B0GCL6 A0A0R1DSM3 A0A0J9TG14 X2J8N3
A0A1B6KZC5 A0A1B6HSG2 A0A194RFI9 A0A1B6D6P0 A0A088A4M7 D2A1I5 A0A1B6GGU9 A0A1B6JD21 A0A0C9PVE1 A0A2P8ZCY9 A0A1B6I016 K7ISA4 A0A195E5N0 E2BLK2 A0A232ENH6 A0A0N0U3F4 A0A151HZ23 A0A0L7R3A0 E2AJP4 F4W4C9 A0A151WXE6 A0A1B6II49 A0A026WJ32 A0A2A3E9C5 T1HK41 U4UMS3 A0A154PQI7 A0A151ILV7 A0A1S4E7E1 U4UN97 A0A067QYJ8 A0A2J7RRW7 A0A1B6M8T7 J9JPK9 E0W319 A0A1I8NVI9 A0A1I8NVJ3 A0A336LY43 A0A0K2T1U8 A0A2S2NAL0 A0A1A9VMJ0 A0A0L0CDH8 B4JDH8 A0A182RDQ9 B4P085 A0A182JF88 B3N4T7 B4N788 A0A182W6G1 A0A182LXR0 A0A310SNH7 A0A1I8NVI7 A0A034W9X2 Q9VMT4 B4Q3J6 A0A0K8TZJ8 A0A182YDU8 A0A084WH79 B4LSN2 A0A1B0AEK7 A0A182L0J9 B4I1E1 A0A182IC70 Q7Q4F7 A0A182XFY3 A0A1W4UV89 Q16YB1 A0A182PLR0 A0A182TRN9 A0A3B0JB05 A0A182NTZ3 A0A182JT46 W8B575 A0A182Q904 B3MLM7 A0A182V911 A0A1S4GYS3 W5JQK2 B4G6P9 Q29M19 J9JU69 B4KG17 A0A0M4E5E9 A0A1A9Y3H1 A0A1Q3G5C7 A0A182G4P9 A0A1J1IJZ5 A0A182F3F6 A0A1B0GCL6 A0A0R1DSM3 A0A0J9TG14 X2J8N3
Pubmed
22118469
26354079
18362917
19820115
29403074
20075255
+ More
20798317 28648823 21719571 24508170 23537049 24845553 20566863 26108605 17994087 17550304 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25244985 24438588 20966253 12364791 17510324 24495485 20920257 23761445 15632085 18057021 26483478
20798317 28648823 21719571 24508170 23537049 24845553 20566863 26108605 17994087 17550304 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25244985 24438588 20966253 12364791 17510324 24495485 20920257 23761445 15632085 18057021 26483478
EMBL
ODYU01011528
SOQ57317.1
RSAL01000023
RVE52269.1
NWSH01000210
PCG78374.1
+ More
AGBW02012542 OWR44853.1 KQ459439 KPJ01041.1 GECZ01002107 JAS67662.1 GEBQ01023207 JAT16770.1 GECU01030110 JAS77596.1 KQ460297 KPJ16224.1 GEDC01015958 JAS21340.1 KQ971338 EFA01534.1 GECZ01008100 JAS61669.1 GECU01010632 JAS97074.1 GBYB01005403 JAG75170.1 PYGN01000098 PSN54369.1 GECU01027446 JAS80260.1 KQ979608 KYN20500.1 GL449033 EFN83439.1 NNAY01003136 OXU19923.1 KQ435885 KOX69582.1 KQ976717 KYM76778.1 KQ414663 KOC65256.1 GL440062 EFN66328.1 GL887513 EGI70942.1 KQ982685 KYQ52341.1 GECU01021115 JAS86591.1 KK107182 EZA56008.1 KZ288315 PBC28320.1 ACPB03017500 KB632373 ERL93793.1 KQ434999 KZC13380.1 KQ977085 KYN05871.1 KB632397 ERL94602.1 KK852872 KDR14568.1 NEVH01000597 PNF43574.1 GEBQ01007637 JAT32340.1 ABLF02027020 ABLF02027024 ABLF02027027 ABLF02027028 ABLF02027030 DS235881 EEB20025.1 UFQS01000219 UFQT01000219 SSX01531.1 SSX21911.1 HACA01002080 CDW19441.1 GGMR01001556 MBY14175.1 JRES01000536 KNC30315.1 CH916368 EDW03348.1 CM000157 EDW88950.1 CH954177 EDV57839.1 CH964182 EDW80229.1 AXCM01000782 KQ760801 OAD58962.1 GAKP01008379 JAC50573.1 AE014134 AY058392 AAF52227.1 AAL13621.1 CM000361 CM002910 EDX03800.1 KMY88264.1 GDHF01032613 JAI19701.1 ATLV01023795 KE525346 KFB49573.1 CH940649 EDW63771.1 CH480820 EDW54348.1 APCN01000778 AAAB01008964 EAA12225.1 CH477519 EAT39615.1 OUUW01000004 SPP79524.1 GAMC01010260 JAB96295.1 AXCN02000807 CH902620 EDV31776.1 ADMH02000622 ETN65578.1 CH479180 EDW29163.1 CH379060 EAL33877.1 ABLF02036331 ABLF02036344 ABLF02036348 ABLF02036353 CH933807 EDW13156.1 KRG03594.1 CP012523 ALC39321.1 GFDL01000035 JAV35010.1 JXUM01144169 KQ569733 KXJ68533.1 CVRI01000054 CRL00555.1 CCAG010021474 KRJ98112.1 KMY88265.1 AHN54131.1
AGBW02012542 OWR44853.1 KQ459439 KPJ01041.1 GECZ01002107 JAS67662.1 GEBQ01023207 JAT16770.1 GECU01030110 JAS77596.1 KQ460297 KPJ16224.1 GEDC01015958 JAS21340.1 KQ971338 EFA01534.1 GECZ01008100 JAS61669.1 GECU01010632 JAS97074.1 GBYB01005403 JAG75170.1 PYGN01000098 PSN54369.1 GECU01027446 JAS80260.1 KQ979608 KYN20500.1 GL449033 EFN83439.1 NNAY01003136 OXU19923.1 KQ435885 KOX69582.1 KQ976717 KYM76778.1 KQ414663 KOC65256.1 GL440062 EFN66328.1 GL887513 EGI70942.1 KQ982685 KYQ52341.1 GECU01021115 JAS86591.1 KK107182 EZA56008.1 KZ288315 PBC28320.1 ACPB03017500 KB632373 ERL93793.1 KQ434999 KZC13380.1 KQ977085 KYN05871.1 KB632397 ERL94602.1 KK852872 KDR14568.1 NEVH01000597 PNF43574.1 GEBQ01007637 JAT32340.1 ABLF02027020 ABLF02027024 ABLF02027027 ABLF02027028 ABLF02027030 DS235881 EEB20025.1 UFQS01000219 UFQT01000219 SSX01531.1 SSX21911.1 HACA01002080 CDW19441.1 GGMR01001556 MBY14175.1 JRES01000536 KNC30315.1 CH916368 EDW03348.1 CM000157 EDW88950.1 CH954177 EDV57839.1 CH964182 EDW80229.1 AXCM01000782 KQ760801 OAD58962.1 GAKP01008379 JAC50573.1 AE014134 AY058392 AAF52227.1 AAL13621.1 CM000361 CM002910 EDX03800.1 KMY88264.1 GDHF01032613 JAI19701.1 ATLV01023795 KE525346 KFB49573.1 CH940649 EDW63771.1 CH480820 EDW54348.1 APCN01000778 AAAB01008964 EAA12225.1 CH477519 EAT39615.1 OUUW01000004 SPP79524.1 GAMC01010260 JAB96295.1 AXCN02000807 CH902620 EDV31776.1 ADMH02000622 ETN65578.1 CH479180 EDW29163.1 CH379060 EAL33877.1 ABLF02036331 ABLF02036344 ABLF02036348 ABLF02036353 CH933807 EDW13156.1 KRG03594.1 CP012523 ALC39321.1 GFDL01000035 JAV35010.1 JXUM01144169 KQ569733 KXJ68533.1 CVRI01000054 CRL00555.1 CCAG010021474 KRJ98112.1 KMY88265.1 AHN54131.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
UP000005203
+ More
UP000007266 UP000245037 UP000002358 UP000078492 UP000008237 UP000215335 UP000053105 UP000078540 UP000053825 UP000000311 UP000007755 UP000075809 UP000053097 UP000242457 UP000015103 UP000030742 UP000076502 UP000078542 UP000079169 UP000027135 UP000235965 UP000007819 UP000009046 UP000095300 UP000078200 UP000037069 UP000001070 UP000075900 UP000002282 UP000075880 UP000008711 UP000007798 UP000075920 UP000075883 UP000000803 UP000000304 UP000076408 UP000030765 UP000008792 UP000092445 UP000075882 UP000001292 UP000075840 UP000007062 UP000076407 UP000192221 UP000008820 UP000075885 UP000075902 UP000268350 UP000075884 UP000075881 UP000075886 UP000007801 UP000075903 UP000000673 UP000008744 UP000001819 UP000009192 UP000092553 UP000092443 UP000069940 UP000249989 UP000183832 UP000069272 UP000092444
UP000007266 UP000245037 UP000002358 UP000078492 UP000008237 UP000215335 UP000053105 UP000078540 UP000053825 UP000000311 UP000007755 UP000075809 UP000053097 UP000242457 UP000015103 UP000030742 UP000076502 UP000078542 UP000079169 UP000027135 UP000235965 UP000007819 UP000009046 UP000095300 UP000078200 UP000037069 UP000001070 UP000075900 UP000002282 UP000075880 UP000008711 UP000007798 UP000075920 UP000075883 UP000000803 UP000000304 UP000076408 UP000030765 UP000008792 UP000092445 UP000075882 UP000001292 UP000075840 UP000007062 UP000076407 UP000192221 UP000008820 UP000075885 UP000075902 UP000268350 UP000075884 UP000075881 UP000075886 UP000007801 UP000075903 UP000000673 UP000008744 UP000001819 UP000009192 UP000092553 UP000092443 UP000069940 UP000249989 UP000183832 UP000069272 UP000092444
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WWF4
A0A437BPH5
A0A2A4K393
A0A212ETN2
A0A194QC78
A0A1B6GZ25
+ More
A0A1B6KZC5 A0A1B6HSG2 A0A194RFI9 A0A1B6D6P0 A0A088A4M7 D2A1I5 A0A1B6GGU9 A0A1B6JD21 A0A0C9PVE1 A0A2P8ZCY9 A0A1B6I016 K7ISA4 A0A195E5N0 E2BLK2 A0A232ENH6 A0A0N0U3F4 A0A151HZ23 A0A0L7R3A0 E2AJP4 F4W4C9 A0A151WXE6 A0A1B6II49 A0A026WJ32 A0A2A3E9C5 T1HK41 U4UMS3 A0A154PQI7 A0A151ILV7 A0A1S4E7E1 U4UN97 A0A067QYJ8 A0A2J7RRW7 A0A1B6M8T7 J9JPK9 E0W319 A0A1I8NVI9 A0A1I8NVJ3 A0A336LY43 A0A0K2T1U8 A0A2S2NAL0 A0A1A9VMJ0 A0A0L0CDH8 B4JDH8 A0A182RDQ9 B4P085 A0A182JF88 B3N4T7 B4N788 A0A182W6G1 A0A182LXR0 A0A310SNH7 A0A1I8NVI7 A0A034W9X2 Q9VMT4 B4Q3J6 A0A0K8TZJ8 A0A182YDU8 A0A084WH79 B4LSN2 A0A1B0AEK7 A0A182L0J9 B4I1E1 A0A182IC70 Q7Q4F7 A0A182XFY3 A0A1W4UV89 Q16YB1 A0A182PLR0 A0A182TRN9 A0A3B0JB05 A0A182NTZ3 A0A182JT46 W8B575 A0A182Q904 B3MLM7 A0A182V911 A0A1S4GYS3 W5JQK2 B4G6P9 Q29M19 J9JU69 B4KG17 A0A0M4E5E9 A0A1A9Y3H1 A0A1Q3G5C7 A0A182G4P9 A0A1J1IJZ5 A0A182F3F6 A0A1B0GCL6 A0A0R1DSM3 A0A0J9TG14 X2J8N3
A0A1B6KZC5 A0A1B6HSG2 A0A194RFI9 A0A1B6D6P0 A0A088A4M7 D2A1I5 A0A1B6GGU9 A0A1B6JD21 A0A0C9PVE1 A0A2P8ZCY9 A0A1B6I016 K7ISA4 A0A195E5N0 E2BLK2 A0A232ENH6 A0A0N0U3F4 A0A151HZ23 A0A0L7R3A0 E2AJP4 F4W4C9 A0A151WXE6 A0A1B6II49 A0A026WJ32 A0A2A3E9C5 T1HK41 U4UMS3 A0A154PQI7 A0A151ILV7 A0A1S4E7E1 U4UN97 A0A067QYJ8 A0A2J7RRW7 A0A1B6M8T7 J9JPK9 E0W319 A0A1I8NVI9 A0A1I8NVJ3 A0A336LY43 A0A0K2T1U8 A0A2S2NAL0 A0A1A9VMJ0 A0A0L0CDH8 B4JDH8 A0A182RDQ9 B4P085 A0A182JF88 B3N4T7 B4N788 A0A182W6G1 A0A182LXR0 A0A310SNH7 A0A1I8NVI7 A0A034W9X2 Q9VMT4 B4Q3J6 A0A0K8TZJ8 A0A182YDU8 A0A084WH79 B4LSN2 A0A1B0AEK7 A0A182L0J9 B4I1E1 A0A182IC70 Q7Q4F7 A0A182XFY3 A0A1W4UV89 Q16YB1 A0A182PLR0 A0A182TRN9 A0A3B0JB05 A0A182NTZ3 A0A182JT46 W8B575 A0A182Q904 B3MLM7 A0A182V911 A0A1S4GYS3 W5JQK2 B4G6P9 Q29M19 J9JU69 B4KG17 A0A0M4E5E9 A0A1A9Y3H1 A0A1Q3G5C7 A0A182G4P9 A0A1J1IJZ5 A0A182F3F6 A0A1B0GCL6 A0A0R1DSM3 A0A0J9TG14 X2J8N3
PDB
2QQ8
E-value=0.0848536,
Score=84
Ontologies
PANTHER
Topology
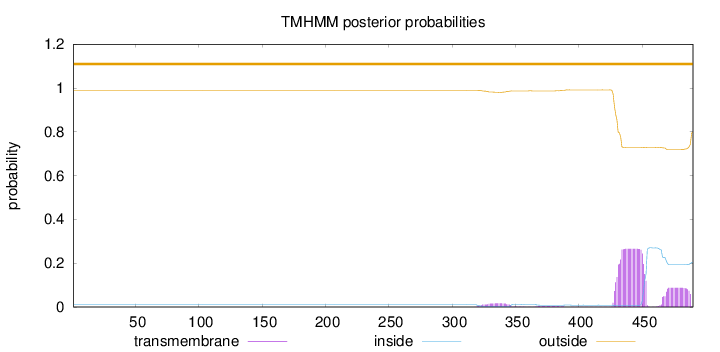
Length:
490
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.22303
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01034
outside
1 - 490
Population Genetic Test Statistics
Pi
2.330089
Theta
20.346948
Tajima's D
-0.777373
CLR
1.576684
CSRT
0.180990950452477
Interpretation
Uncertain
