Gene
KWMTBOMO08108
Pre Gene Modal
BGIBMGA001061
Annotation
PREDICTED:_alkylated_DNA_repair_protein_alkB_homolog_8_[Plutella_xylostella]
Full name
Alkylated DNA repair protein alkB homolog 8
Alternative Name
Probable alpha-ketoglutarate-dependent dioxygenase ABH8
S-adenosyl-L-methionine-dependent tRNA methyltransferase ABH8
tRNA (carboxymethyluridine(34)-5-O)-methyltransferase ABH8
S-adenosyl-L-methionine-dependent tRNA methyltransferase ABH8
tRNA (carboxymethyluridine(34)-5-O)-methyltransferase ABH8
Location in the cell
Cytoplasmic Reliability : 1.795 Nuclear Reliability : 1.931
Sequence
CDS
ATGGAAGACGATAGAAAACATGAGAAGAAACGCAAACGGTTTGCTGCCCGATTGAATACTCTGAAAGGAATTGTTTGTTATGAAGATCCTAGGCCCAATGTGATTCTTTGCAATGTGGGGCAAGCTACAGGATTTGATAAGCAAAAACTGTTGAAACTGCTCACAGAGTACAAGGTAAACACACCATTGCCTAAGTTTGTTGCCGAGAAAGGAGAATCCTACTCATATCTCATGTTTGATAAAGCCGAGAATGCATCACTATTTTATGAGGCATGCAATGGGAAAGCACAGGTAGATGAGAATGGAACAACACTCTATGTGACATTTGTTGAAAATGTTCCGGATACTGATATTATTTGTAAGCATCCAAATCCTAAAGGCTTACATATAATACCCGATTTTCTTACCGAAGATGAAGAGAAGTTATTCCTTAATACCTTTAACATACAACACGTAGAAACAACATTAAAGAACCGTCAAGTCAAACATTATGGTTATGAGTTCAGATATGGCAGCAATGACGTTGACTTGAGTTGCCCATTGCAGGAAAAAATACCAAAGATTTGTAAATTGCTGTGGAGAAGGCTCCAACACTATGGTTACGATCTCGGAGTACCAGTTCAGCTGACTGTTAACAAGTACTTACCTGGGCAAGGTATCCCGTCGCATGTCGACAAACACAGTCCATTTGGAGAAACAATTCTGGCCCTATCACTGGGCTCCAATGTGGTTATGGATTGGAAGCATCACACAGGGAAATATGTGCCAGTTATGGTTGAGGCTAGATCAATGATGATAATGCAAGATGAAGCTAGATATGACTGGCAGCATGGAATACAGCCAAGAATGTGGGACCCAGTCCTTGATGTTCGTTTAATAGATAATGAGAAAGTTAAAGTGATCACATCTGATACTGTACAGAGGGAGATGAGAATCTCCTTGACATTCCGATGGACCAGATGCGGGCCATGTAAATGTGAATACAAGATGTTGTGTGATAGCATTGAACGTAATATTCCTGAGACAATTAGCAATGATGTTGCATCTAACATAGAGGACATACATGTGCACCAGGTATACGAACAAATCGCAGGACACTTCAGCACAACGCGGCACAAGCCTTGGCCCAAAGTTGTAGACTTCATGCAGCACGTGCCGGCTGGATCTATAGTGCTGGACCTTGGCTGTGGCAATGGCAAGAATATACTCAACAGAACAGATATATTGCAGGTAGCCGGTGAAAGGAGCAGCGGTCTACTAGAGGAATGCAAGGGGCTCACGGCCCGGATATCCGGCGCCGACTGCATCCGGTTGGACTTGCTCAACACCGGCCTCAAGGACGAGTGCGCTGATTTCATCATTTGCGTAGCCGTCGTGCACCACTTCAGCACACAGGCACGGCGCCTCCACTCGCTACAAACAATACACCGACTCCTGCGGACCAACGGGCAAGCGCTCATCACGGTGTGGGCCAAGGATCAAACCAAGTCGAGCTACCTGTCCCGGACCCGGGCGCCGCTGCTGGACCGGCACAAACTGACGGTGGGCGGCATCCACTTGCCCGTGCACGAGAACCGCACGCAGTTCCAGCACAAGGATCTGCTGGTGCCCTGGAACCTGCGCAGCAAGGCGCCCCCGCTCCAACAGCCCGACACCACCTTTCTCAGATACTACCACGTCTTCGATGAAGGGGAACTGGATCAGCTCTGCCGAGACGCGCGGCTCGCGATCGTCAGGAGCTTCTACGAGGAAGGCAATTGGTGTGTCGTCTGTATGAAAGTGTAA
Protein
MEDDRKHEKKRKRFAARLNTLKGIVCYEDPRPNVILCNVGQATGFDKQKLLKLLTEYKVNTPLPKFVAEKGESYSYLMFDKAENASLFYEACNGKAQVDENGTTLYVTFVENVPDTDIICKHPNPKGLHIIPDFLTEDEEKLFLNTFNIQHVETTLKNRQVKHYGYEFRYGSNDVDLSCPLQEKIPKICKLLWRRLQHYGYDLGVPVQLTVNKYLPGQGIPSHVDKHSPFGETILALSLGSNVVMDWKHHTGKYVPVMVEARSMMIMQDEARYDWQHGIQPRMWDPVLDVRLIDNEKVKVITSDTVQREMRISLTFRWTRCGPCKCEYKMLCDSIERNIPETISNDVASNIEDIHVHQVYEQIAGHFSTTRHKPWPKVVDFMQHVPAGSIVLDLGCGNGKNILNRTDILQVAGERSSGLLEECKGLTARISGADCIRLDLLNTGLKDECADFIICVAVVHHFSTQARRLHSLQTIHRLLRTNGQALITVWAKDQTKSSYLSRTRAPLLDRHKLTVGGIHLPVHENRTQFQHKDLLVPWNLRSKAPPLQQPDTTFLRYYHVFDEGELDQLCRDARLAIVRSFYEEGNWCVVCMKV
Summary
Description
Catalyzes the methylation of 5-carboxymethyl uridine to 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its methyltransferase domain. Catalyzes the last step in the formation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in target tRNA. Has a preference for tRNA(Arg) and tRNA(Glu), and does not bind tRNA(Lys). Binds tRNA and catalyzes the iron and alpha-ketoglutarate dependent hydroxylation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its dioxygenase domain, giving rise to 5-(S)-methoxycarbonylhydroxymethyluridine; has a preference for tRNA(Gly). Required for normal survival after DNA damage. May inhibit apoptosis and promote cell survival and angiogenesis (By similarity).
Catalytic Activity
carboxymethyluridine(34) in tRNA + S-adenosyl-L-methionine = 5-(2-methoxy-2-oxoethyl)uridine(34) in tRNA + S-adenosyl-L-homocysteine
Cofactor
Fe(2+)
Similarity
Belongs to the alkB family.
Keywords
Complete proteome
Cytoplasm
Iron
Metal-binding
Methyltransferase
Multifunctional enzyme
Nucleus
Oxidoreductase
Reference proteome
RNA-binding
S-adenosyl-L-methionine
Transferase
Zinc
Feature
chain Alkylated DNA repair protein alkB homolog 8
Uniprot
H9IUY1
A0A2A4K3N2
A0A437BPC4
A0A2A4K3Z1
A0A194Q7R7
A0A212ETM0
+ More
A0A194REG2 A0A1A9ZNI5 A0A1Y1N078 A0A1A9VIH1 A0A2J7QS63 A0A2Z5U609 A0A1B0D934 A0A0A1X0P0 D2A2C2 A0A1W4WS58 A0A1B0BGD9 Q17MR7 A0A182Y9F9 A0A1A9XJ53 A0A2P8ZL88 A0A084WIN4 A0A1I8NHX9 A0A0T6AXC8 A0A067R5V9 A0A0N0BEQ0 W5JWS8 A0A182IX61 A0A2M4CLJ7 A0A182RS07 A0A1B6E4R5 A0A182PYF3 A0A182M3B7 A0A182N2V9 A0A2R7WGB7 A0A336MWL3 E2B3I8 A0A2M4BHG6 A0A2M4BH89 W8BCH1 Q7PZG8 A0A182L530 A0A1S4H7U0 K7J2T9 A0A182XJT8 A0A182UZ32 A0A336M1J7 A0A182TKL7 A0A182QSX5 A0A0L0C1L5 Q9W232 A0A1W4VQN5 Q290H5 N6TXH5 F6ZD81 Q8T9A3 A0A0L7RKL6 A0A088A8Y1 A0A2A3E5W1 B4GC19 B4I868 U4U8C2 B4QHJ3 Q07G10 B4P5H7 A0A096N913 A0A2K5Y7X6 A0A096NCH5 A0A1S3W968 A0A0M5J527 B3NLI7 A0A1S2ZKC4 G1T292 A0A154PK51 K9KC91 F7DYZ5 A0A2I2US18 E2AE04 A0A182KIK7 A0A182WDY8 A0A3B0J0S8 A0A232EJC4 A0A1I8PW84 A0A337S599 A0A3Q7TMR0 E2REE7 A0A3Q7SL51 G3WMU7 A0A182HHJ4 T1J3X4 A0A384C0E5 A0A2Y9DXP8 A0A3Q7WJ91 A0A452TTN1 B3MEC7 E9IFM0 A0A3Q7X508 A0A3Q7XGG4 A0A2K5DGZ9 U3B9R7
A0A194REG2 A0A1A9ZNI5 A0A1Y1N078 A0A1A9VIH1 A0A2J7QS63 A0A2Z5U609 A0A1B0D934 A0A0A1X0P0 D2A2C2 A0A1W4WS58 A0A1B0BGD9 Q17MR7 A0A182Y9F9 A0A1A9XJ53 A0A2P8ZL88 A0A084WIN4 A0A1I8NHX9 A0A0T6AXC8 A0A067R5V9 A0A0N0BEQ0 W5JWS8 A0A182IX61 A0A2M4CLJ7 A0A182RS07 A0A1B6E4R5 A0A182PYF3 A0A182M3B7 A0A182N2V9 A0A2R7WGB7 A0A336MWL3 E2B3I8 A0A2M4BHG6 A0A2M4BH89 W8BCH1 Q7PZG8 A0A182L530 A0A1S4H7U0 K7J2T9 A0A182XJT8 A0A182UZ32 A0A336M1J7 A0A182TKL7 A0A182QSX5 A0A0L0C1L5 Q9W232 A0A1W4VQN5 Q290H5 N6TXH5 F6ZD81 Q8T9A3 A0A0L7RKL6 A0A088A8Y1 A0A2A3E5W1 B4GC19 B4I868 U4U8C2 B4QHJ3 Q07G10 B4P5H7 A0A096N913 A0A2K5Y7X6 A0A096NCH5 A0A1S3W968 A0A0M5J527 B3NLI7 A0A1S2ZKC4 G1T292 A0A154PK51 K9KC91 F7DYZ5 A0A2I2US18 E2AE04 A0A182KIK7 A0A182WDY8 A0A3B0J0S8 A0A232EJC4 A0A1I8PW84 A0A337S599 A0A3Q7TMR0 E2REE7 A0A3Q7SL51 G3WMU7 A0A182HHJ4 T1J3X4 A0A384C0E5 A0A2Y9DXP8 A0A3Q7WJ91 A0A452TTN1 B3MEC7 E9IFM0 A0A3Q7X508 A0A3Q7XGG4 A0A2K5DGZ9 U3B9R7
EC Number
1.14.11.-
Pubmed
19121390
26354079
22118469
28004739
26760975
25830018
+ More
18362917 19820115 17510324 25244985 29403074 24438588 25315136 24845553 20920257 23761445 20798317 24495485 12364791 20966253 20075255 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17994087 23537049 17495919 22936249 17550304 21993624 19892987 17975172 28648823 16341006 21709235 24813606 21282665 25243066
18362917 19820115 17510324 25244985 29403074 24438588 25315136 24845553 20920257 23761445 20798317 24495485 12364791 20966253 20075255 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17994087 23537049 17495919 22936249 17550304 21993624 19892987 17975172 28648823 16341006 21709235 24813606 21282665 25243066
EMBL
BABH01000167
NWSH01000210
PCG78373.1
RSAL01000023
RVE52268.1
PCG78372.1
+ More
KQ459439 KPJ01040.1 AGBW02012542 OWR44852.1 KQ460297 KPJ16223.1 GEZM01019409 JAV89646.1 NEVH01011878 PNF31428.1 FX985781 BBA93668.1 AJVK01012889 GBXI01009992 JAD04300.1 KQ971338 EFA02043.1 JXJN01013833 CH477204 EAT47955.1 PYGN01000024 PSN57265.1 ATLV01023941 KE525347 KFB50078.1 LJIG01022609 KRT79652.1 KK852866 KDR14734.1 KQ435827 KOX71948.1 ADMH02000120 ETN67745.1 GGFL01002032 MBW66210.1 GEDC01004375 JAS32923.1 AXCM01000907 KK854778 PTY18646.1 UFQT01002849 SSX34135.1 GL445346 EFN89740.1 GGFJ01003260 MBW52401.1 GGFJ01003261 MBW52402.1 GAMC01011852 JAB94703.1 AAAB01008986 EAA00113.2 UFQT01000416 SSX24115.1 AXCN02001090 JRES01001012 KNC26152.1 AE013599 AAF46867.1 CM000071 EAL25387.2 APGK01051339 KB741189 ENN73061.1 AY069858 AAL40003.1 KQ414573 KOC71296.1 KZ288379 PBC26559.1 CH479181 EDW32362.1 CH480824 EDW56793.1 KB632216 ERL90169.1 CM000362 CM002911 EDX08222.1 KMY95817.1 CR855607 CM000158 EDW91808.1 AHZZ02007567 AHZZ02007568 AHZZ02007569 AHZZ02007570 AHZZ02007571 AHZZ02007572 CP012524 ALC42006.1 CH954179 EDV54903.1 AAGW02009189 AAGW02009190 AAGW02009191 KQ434943 KZC12222.1 JL620619 AEP99344.1 AANG04000369 GL438827 EFN68237.1 OUUW01000001 SPP74337.1 NNAY01004036 OXU18466.1 AAEX03003519 AEFK01115055 AEFK01115056 AEFK01115057 AEFK01115058 AEFK01115059 APCN01002417 JH431832 CH902619 EDV36533.1 GL762865 EFZ20637.1 GAMT01009532 JAB02329.1
KQ459439 KPJ01040.1 AGBW02012542 OWR44852.1 KQ460297 KPJ16223.1 GEZM01019409 JAV89646.1 NEVH01011878 PNF31428.1 FX985781 BBA93668.1 AJVK01012889 GBXI01009992 JAD04300.1 KQ971338 EFA02043.1 JXJN01013833 CH477204 EAT47955.1 PYGN01000024 PSN57265.1 ATLV01023941 KE525347 KFB50078.1 LJIG01022609 KRT79652.1 KK852866 KDR14734.1 KQ435827 KOX71948.1 ADMH02000120 ETN67745.1 GGFL01002032 MBW66210.1 GEDC01004375 JAS32923.1 AXCM01000907 KK854778 PTY18646.1 UFQT01002849 SSX34135.1 GL445346 EFN89740.1 GGFJ01003260 MBW52401.1 GGFJ01003261 MBW52402.1 GAMC01011852 JAB94703.1 AAAB01008986 EAA00113.2 UFQT01000416 SSX24115.1 AXCN02001090 JRES01001012 KNC26152.1 AE013599 AAF46867.1 CM000071 EAL25387.2 APGK01051339 KB741189 ENN73061.1 AY069858 AAL40003.1 KQ414573 KOC71296.1 KZ288379 PBC26559.1 CH479181 EDW32362.1 CH480824 EDW56793.1 KB632216 ERL90169.1 CM000362 CM002911 EDX08222.1 KMY95817.1 CR855607 CM000158 EDW91808.1 AHZZ02007567 AHZZ02007568 AHZZ02007569 AHZZ02007570 AHZZ02007571 AHZZ02007572 CP012524 ALC42006.1 CH954179 EDV54903.1 AAGW02009189 AAGW02009190 AAGW02009191 KQ434943 KZC12222.1 JL620619 AEP99344.1 AANG04000369 GL438827 EFN68237.1 OUUW01000001 SPP74337.1 NNAY01004036 OXU18466.1 AAEX03003519 AEFK01115055 AEFK01115056 AEFK01115057 AEFK01115058 AEFK01115059 APCN01002417 JH431832 CH902619 EDV36533.1 GL762865 EFZ20637.1 GAMT01009532 JAB02329.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000092445 UP000078200 UP000235965 UP000092462 UP000007266 UP000192223 UP000092460 UP000008820 UP000076408 UP000092443 UP000245037 UP000030765 UP000095301 UP000027135 UP000053105 UP000000673 UP000075880 UP000075900 UP000075885 UP000075883 UP000075884 UP000008237 UP000007062 UP000075882 UP000002358 UP000076407 UP000075903 UP000075902 UP000075886 UP000037069 UP000000803 UP000192221 UP000001819 UP000019118 UP000002280 UP000053825 UP000005203 UP000242457 UP000008744 UP000001292 UP000030742 UP000000304 UP000008143 UP000002282 UP000028761 UP000233140 UP000079721 UP000092553 UP000008711 UP000001811 UP000076502 UP000002281 UP000011712 UP000000311 UP000075881 UP000075920 UP000268350 UP000215335 UP000095300 UP000286640 UP000002254 UP000007648 UP000075840 UP000261680 UP000291021 UP000248480 UP000286642 UP000007801 UP000233020
UP000092445 UP000078200 UP000235965 UP000092462 UP000007266 UP000192223 UP000092460 UP000008820 UP000076408 UP000092443 UP000245037 UP000030765 UP000095301 UP000027135 UP000053105 UP000000673 UP000075880 UP000075900 UP000075885 UP000075883 UP000075884 UP000008237 UP000007062 UP000075882 UP000002358 UP000076407 UP000075903 UP000075902 UP000075886 UP000037069 UP000000803 UP000192221 UP000001819 UP000019118 UP000002280 UP000053825 UP000005203 UP000242457 UP000008744 UP000001292 UP000030742 UP000000304 UP000008143 UP000002282 UP000028761 UP000233140 UP000079721 UP000092553 UP000008711 UP000001811 UP000076502 UP000002281 UP000011712 UP000000311 UP000075881 UP000075920 UP000268350 UP000215335 UP000095300 UP000286640 UP000002254 UP000007648 UP000075840 UP000261680 UP000291021 UP000248480 UP000286642 UP000007801 UP000233020
Pfam
Interpro
IPR005123
Oxoglu/Fe-dep_dioxygenase
+ More
IPR013216 Methyltransf_11
IPR027450 AlkB-like
IPR029063 SAM-dependent_MTases
IPR032863 ALKBH8
IPR037151 AlkB-like_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR034256 ALKBH8_RRM
IPR041698 Methyltransf_25
IPR015095 AlkB_hom8_N
IPR013216 Methyltransf_11
IPR027450 AlkB-like
IPR029063 SAM-dependent_MTases
IPR032863 ALKBH8
IPR037151 AlkB-like_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR034256 ALKBH8_RRM
IPR041698 Methyltransf_25
IPR015095 AlkB_hom8_N
Gene 3D
ProteinModelPortal
H9IUY1
A0A2A4K3N2
A0A437BPC4
A0A2A4K3Z1
A0A194Q7R7
A0A212ETM0
+ More
A0A194REG2 A0A1A9ZNI5 A0A1Y1N078 A0A1A9VIH1 A0A2J7QS63 A0A2Z5U609 A0A1B0D934 A0A0A1X0P0 D2A2C2 A0A1W4WS58 A0A1B0BGD9 Q17MR7 A0A182Y9F9 A0A1A9XJ53 A0A2P8ZL88 A0A084WIN4 A0A1I8NHX9 A0A0T6AXC8 A0A067R5V9 A0A0N0BEQ0 W5JWS8 A0A182IX61 A0A2M4CLJ7 A0A182RS07 A0A1B6E4R5 A0A182PYF3 A0A182M3B7 A0A182N2V9 A0A2R7WGB7 A0A336MWL3 E2B3I8 A0A2M4BHG6 A0A2M4BH89 W8BCH1 Q7PZG8 A0A182L530 A0A1S4H7U0 K7J2T9 A0A182XJT8 A0A182UZ32 A0A336M1J7 A0A182TKL7 A0A182QSX5 A0A0L0C1L5 Q9W232 A0A1W4VQN5 Q290H5 N6TXH5 F6ZD81 Q8T9A3 A0A0L7RKL6 A0A088A8Y1 A0A2A3E5W1 B4GC19 B4I868 U4U8C2 B4QHJ3 Q07G10 B4P5H7 A0A096N913 A0A2K5Y7X6 A0A096NCH5 A0A1S3W968 A0A0M5J527 B3NLI7 A0A1S2ZKC4 G1T292 A0A154PK51 K9KC91 F7DYZ5 A0A2I2US18 E2AE04 A0A182KIK7 A0A182WDY8 A0A3B0J0S8 A0A232EJC4 A0A1I8PW84 A0A337S599 A0A3Q7TMR0 E2REE7 A0A3Q7SL51 G3WMU7 A0A182HHJ4 T1J3X4 A0A384C0E5 A0A2Y9DXP8 A0A3Q7WJ91 A0A452TTN1 B3MEC7 E9IFM0 A0A3Q7X508 A0A3Q7XGG4 A0A2K5DGZ9 U3B9R7
A0A194REG2 A0A1A9ZNI5 A0A1Y1N078 A0A1A9VIH1 A0A2J7QS63 A0A2Z5U609 A0A1B0D934 A0A0A1X0P0 D2A2C2 A0A1W4WS58 A0A1B0BGD9 Q17MR7 A0A182Y9F9 A0A1A9XJ53 A0A2P8ZL88 A0A084WIN4 A0A1I8NHX9 A0A0T6AXC8 A0A067R5V9 A0A0N0BEQ0 W5JWS8 A0A182IX61 A0A2M4CLJ7 A0A182RS07 A0A1B6E4R5 A0A182PYF3 A0A182M3B7 A0A182N2V9 A0A2R7WGB7 A0A336MWL3 E2B3I8 A0A2M4BHG6 A0A2M4BH89 W8BCH1 Q7PZG8 A0A182L530 A0A1S4H7U0 K7J2T9 A0A182XJT8 A0A182UZ32 A0A336M1J7 A0A182TKL7 A0A182QSX5 A0A0L0C1L5 Q9W232 A0A1W4VQN5 Q290H5 N6TXH5 F6ZD81 Q8T9A3 A0A0L7RKL6 A0A088A8Y1 A0A2A3E5W1 B4GC19 B4I868 U4U8C2 B4QHJ3 Q07G10 B4P5H7 A0A096N913 A0A2K5Y7X6 A0A096NCH5 A0A1S3W968 A0A0M5J527 B3NLI7 A0A1S2ZKC4 G1T292 A0A154PK51 K9KC91 F7DYZ5 A0A2I2US18 E2AE04 A0A182KIK7 A0A182WDY8 A0A3B0J0S8 A0A232EJC4 A0A1I8PW84 A0A337S599 A0A3Q7TMR0 E2REE7 A0A3Q7SL51 G3WMU7 A0A182HHJ4 T1J3X4 A0A384C0E5 A0A2Y9DXP8 A0A3Q7WJ91 A0A452TTN1 B3MEC7 E9IFM0 A0A3Q7X508 A0A3Q7XGG4 A0A2K5DGZ9 U3B9R7
PDB
3THT
E-value=6.67637e-49,
Score=491
Ontologies
GO
PANTHER
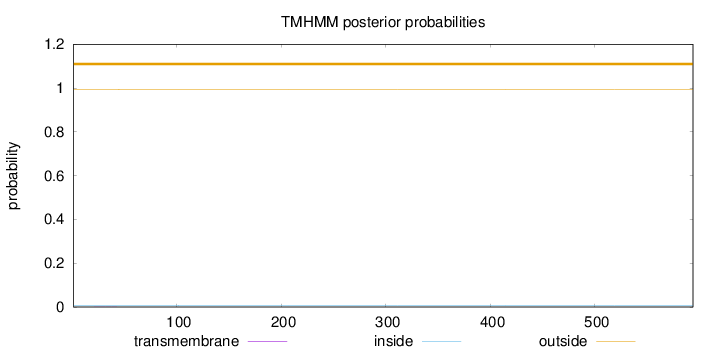
Topology
Subcellular location
Cytoplasm
Predominantly cytoplasmic. With evidence from 2 publications.
Nucleus Predominantly cytoplasmic. With evidence from 2 publications.
Nucleus Predominantly cytoplasmic. With evidence from 2 publications.
Length:
594
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00479999999999999
Exp number, first 60 AAs:
0.00342
Total prob of N-in:
0.00622
outside
1 - 594
Population Genetic Test Statistics
Pi
14.139163
Theta
14.992233
Tajima's D
-0.434835
CLR
0.338162
CSRT
0.260786960651967
Interpretation
Uncertain
