Gene
KWMTBOMO08107
Pre Gene Modal
BGIBMGA001102
Annotation
PREDICTED:_phosphatidylinositol_N-acetylglucosaminyltransferase_subunit_A_[Papilio_polytes]
Full name
Tryptophan 2,3-dioxygenase
Alternative Name
Tryptamin 2,3-dioxygenase
Tryptophan oxygenase
Tryptophan pyrrolase
Tryptophanase
Tryptophan oxygenase
Tryptophan pyrrolase
Tryptophanase
Location in the cell
PlasmaMembrane Reliability : 3.669
Sequence
CDS
ATGGATACTAGAAAAAAGAGGCAAGTTGTTTGTATGGCTTCAGATTTCTTTTACCCGAATACCGGAGGTGTAGAAGAGCATATATTTAATTTGTCACAATGTCTCATTAAACGAGGCCACAAAGTCATAGTACTAACTCACTCTTATGGCGACAGAGTCGGTATCCGATATTTAACCGCAGGCTTAAAAGTTTACTATTTACCTATTAGAGTATTTTATGCCCAGTGCGTATTGCCAACAATGATATGTAATATAGCTTTAATAAGAAATATATTGATAAGAGAATGCATTGAAATAGTTCATGGTCATTCTGCTTTTAGTGTTTTGTGTCATGAAGTATGTATTATAGGCAAATTAATGGGATTAAAAACTGTATTCACTGACCACAGTTTATTTGGGTTTGCTGACACATCAGCTGTACTGACAAACAAGTATTTACAAATATGTCTCAGTGAGATTGATCATTGCATATGTGTATCTCACACTGGCAAAGAGAACACAGTGTTAAGGGCCAAAGTTCAAGCTCACAAAGTTTCGGTGATCCCAAATGCAGTTGATGCATTTTCTTTCATTCCTGATCCATGCCAAAGAGATCAAAAATTTATAACCATAGTCATAGTCTCAAGATTAGTGTACCGAAAAGGTGTTAATCTCATGGCTGCCGTTATAGCAGATATGTGCCCACGGTATCCAAACTTAAGATTCATCATAGGTGGAGATGGTCCTAAGATGTGGCTACTACAAGAAGTCCGAGAAAAAATAGGTTGTCAAGAATCTGTTAGATTGTTAGGCAGCTTAAAACATTCAGAAGTTAGAAATGTTTTAGTGAAGGGTGACATTTTTTTAAATACATCACTGACAGAAGCATATTGTATGGCTATAGTAGAAGCTGCAGCTTGTGGACTAAAAGTTGTTTCTACAAAAGTAGGAGGAATACCTGAAGTACTACCAGAAAGTATGATCTATCTTACGGAGCCAAATGTAGGCAGCCTGGTCAGAGGAATTGAGAAAGCAATAACTGATATTAAAGAAGGAAACATAATGTGTCCATTTAAATGCAATAGACTGGTTCGAGAGATGTACAGTTGGATGGACATTACAAAAAGAACTGAGATAGTTTATGATAGAATATTACTAAACAAAAATAAACCACTAGGGCAACAGTTACGTAGCTACCTTAATTCTGGTGTTTGGCCCTTTCTATTAGTGATAAGTTTAATGTATATATTACTACAATTAACTGAGAGAATTTATAAAAGGAAGTATATTGACAAAGCAAGAGACTTAAGACTATAA
Protein
MDTRKKRQVVCMASDFFYPNTGGVEEHIFNLSQCLIKRGHKVIVLTHSYGDRVGIRYLTAGLKVYYLPIRVFYAQCVLPTMICNIALIRNILIRECIEIVHGHSAFSVLCHEVCIIGKLMGLKTVFTDHSLFGFADTSAVLTNKYLQICLSEIDHCICVSHTGKENTVLRAKVQAHKVSVIPNAVDAFSFIPDPCQRDQKFITIVIVSRLVYRKGVNLMAAVIADMCPRYPNLRFIIGGDGPKMWLLQEVREKIGCQESVRLLGSLKHSEVRNVLVKGDIFLNTSLTEAYCMAIVEAAACGLKVVSTKVGGIPEVLPESMIYLTEPNVGSLVRGIEKAITDIKEGNIMCPFKCNRLVREMYSWMDITKRTEIVYDRILLNKNKPLGQQLRSYLNSGVWPFLLVISLMYILLQLTERIYKRKYIDKARDLRL
Summary
Description
Heme-dependent dioxygenase that catalyzes the oxidative cleavage of the L-tryptophan (L-Trp) pyrrole ring and converts L-tryptophan to N-formyl-L-kynurenine. Catalyzes the oxidative cleavage of the indole moiety.
Catalytic Activity
L-tryptophan + O2 = N-formyl-L-kynurenine
Cofactor
heme
Subunit
Homotetramer. Dimer of dimers.
Similarity
Belongs to the tryptophan 2,3-dioxygenase family.
Uniprot
H9IV22
A0A3S2NNN1
A0A0L7KQ62
A0A0L7KLE8
A0A0L7K3A9
S4PBW5
+ More
A0A212ETS1 A0A2P8YCF1 A0A087ZSH3 A0A0L7RA36 A0A0M8ZT40 A0A2A3ED53 K7JCX9 A0A067R1I8 A0A3L8DSA7 D6WMV9 E2BGW2 A0A310SIC5 A0A232F677 E9JCK3 A0A158NHG6 A0A195AV25 A0A1B6GWA2 A0A1B6JHI6 E2AJJ6 A0A0J7KGJ8 A0A195D4E7 N6UDH8 U4UXZ2 J3JVX8 A0A195FWR3 A0A1B6MT62 A0A154PBE8 F4WDB6 A0A2J7QVI0 A0A151XFF3 A0A195E2X2 A0A2S2QLV1 A0A1B6D3B5 A0A2S2NLC4 J9JRN9 A0A1Y1K0N5 A0A1W4X9R6 A0A023F415 A0A1Q3G1F4 T1HF81 A0A1W4XK61 A0A1Q3G1F2 A0A1L8E3Z1 E0VJ33 A0A1B0FY58 Q179Y5 A0A1S4FAQ9 A0A2H1WW88 A0A1Y1K6K6 A0A182Q6V2 A0A182JLZ3 A0A182YIV0 A0A182PWE4 A0A182NV45 A0A182HYB5 A0A182FVE9 W5JC44 A0A182REL8 A0A1S4H781 A0A182WYS0 A0A182UMW5 A0A182TF14 A0A0P4YYC7 A0A0P5C6U4 A0A0A9YY67 A0A1I8PX39 A0A182H9D4 A0A0P5CHJ2 A0A0P5FBW7 A0A0P4XWI3 A0A182MEU3 E9FRT0 A0A0P6DK33 A0A1J1HXX0 A0A182JSR7 A0A182W9M6 A0A1Y1NEH3 B0WDE0 A0A0P4ZRJ3 A0A0L8G4J0 A0A0P6FP95 A0A210PV17 A0A3R7PWR9 Q7QHN7 C3ZXB6 A0A182SMQ4 A0A1Z5LFC9 A0A1X0RWD4 A0A367JXR2 A0A367JQ78 A0A168PLQ4 A0A2G4TA98
A0A212ETS1 A0A2P8YCF1 A0A087ZSH3 A0A0L7RA36 A0A0M8ZT40 A0A2A3ED53 K7JCX9 A0A067R1I8 A0A3L8DSA7 D6WMV9 E2BGW2 A0A310SIC5 A0A232F677 E9JCK3 A0A158NHG6 A0A195AV25 A0A1B6GWA2 A0A1B6JHI6 E2AJJ6 A0A0J7KGJ8 A0A195D4E7 N6UDH8 U4UXZ2 J3JVX8 A0A195FWR3 A0A1B6MT62 A0A154PBE8 F4WDB6 A0A2J7QVI0 A0A151XFF3 A0A195E2X2 A0A2S2QLV1 A0A1B6D3B5 A0A2S2NLC4 J9JRN9 A0A1Y1K0N5 A0A1W4X9R6 A0A023F415 A0A1Q3G1F4 T1HF81 A0A1W4XK61 A0A1Q3G1F2 A0A1L8E3Z1 E0VJ33 A0A1B0FY58 Q179Y5 A0A1S4FAQ9 A0A2H1WW88 A0A1Y1K6K6 A0A182Q6V2 A0A182JLZ3 A0A182YIV0 A0A182PWE4 A0A182NV45 A0A182HYB5 A0A182FVE9 W5JC44 A0A182REL8 A0A1S4H781 A0A182WYS0 A0A182UMW5 A0A182TF14 A0A0P4YYC7 A0A0P5C6U4 A0A0A9YY67 A0A1I8PX39 A0A182H9D4 A0A0P5CHJ2 A0A0P5FBW7 A0A0P4XWI3 A0A182MEU3 E9FRT0 A0A0P6DK33 A0A1J1HXX0 A0A182JSR7 A0A182W9M6 A0A1Y1NEH3 B0WDE0 A0A0P4ZRJ3 A0A0L8G4J0 A0A0P6FP95 A0A210PV17 A0A3R7PWR9 Q7QHN7 C3ZXB6 A0A182SMQ4 A0A1Z5LFC9 A0A1X0RWD4 A0A367JXR2 A0A367JQ78 A0A168PLQ4 A0A2G4TA98
EC Number
1.13.11.11
Pubmed
19121390
26227816
23622113
22118469
29403074
20075255
+ More
24845553 30249741 18362917 19820115 20798317 28648823 21282665 21347285 23537049 22516182 21719571 28004739 25474469 20566863 17510324 25244985 20920257 23761445 12364791 25401762 26823975 26483478 21292972 28812685 18563158 28528879 27956601 29674435
24845553 30249741 18362917 19820115 20798317 28648823 21282665 21347285 23537049 22516182 21719571 28004739 25474469 20566863 17510324 25244985 20920257 23761445 12364791 25401762 26823975 26483478 21292972 28812685 18563158 28528879 27956601 29674435
EMBL
BABH01000168
RSAL01000023
RVE52267.1
JTDY01007400
KOB65235.1
JTDY01009065
+ More
JTDY01003664 KOB64137.1 KOB69190.1 JTDY01012558 KOB52256.1 GAIX01004416 JAA88144.1 AGBW02012542 OWR44851.1 PYGN01000705 PSN41939.1 KQ414619 KOC67725.1 KQ435903 KOX69083.1 KZ288280 PBC29640.1 AAZX01005171 KK853097 KDR11448.1 QOIP01000005 RLU22739.1 KQ971343 EFA04308.1 GL448228 EFN85094.1 KQ760974 OAD58574.1 NNAY01000869 OXU26135.1 GL771844 EFZ09450.1 ADTU01015804 ADTU01015805 ADTU01015806 KQ976737 KYM75907.1 GECZ01003136 JAS66633.1 GECU01008986 JAS98720.1 GL440027 EFN66391.1 LBMM01007854 KMQ89369.1 KQ976870 KYN07752.1 APGK01032712 KB740848 ENN78681.1 KB632411 ERL95236.1 BT127396 AEE62358.1 KQ981208 KYN44878.1 GEBQ01000889 JAT39088.1 KQ434868 KZC09171.1 GL888086 EGI67843.1 NEVH01010475 PNF32591.1 KQ982194 KYQ59065.1 KQ979763 KYN19234.1 GGMS01009546 MBY78749.1 GEDC01017130 JAS20168.1 GGMR01005374 MBY17993.1 ABLF02014276 GEZM01096214 JAV55049.1 GBBI01002521 JAC16191.1 GFDL01001418 JAV33627.1 ACPB03010315 GFDL01001413 JAV33632.1 GFDF01000829 JAV13255.1 DS235219 EEB13389.1 AJVK01031256 AJVK01031257 CH477343 EAT43050.1 ODYU01011528 SOQ57318.1 GEZM01096216 JAV55046.1 AXCN02001805 APCN01004514 ADMH02001629 ETN61696.1 AAAB01008816 GDIP01229894 GDIP01221186 GDIP01161345 GDIP01154999 JAJ02216.1 GDIP01175352 JAJ48050.1 GBHO01022343 GBHO01006490 GDHC01012304 JAG21261.1 JAG37114.1 JAQ06325.1 JXUM01120466 KQ566395 KXJ70160.1 GDIP01175351 JAJ48051.1 GDIQ01257320 JAJ94404.1 GDIP01245655 JAI77746.1 AXCM01003607 GL732523 EFX89900.1 GDIQ01076554 JAN18183.1 CVRI01000027 CRK92402.1 GEZM01004857 JAV96292.1 DS231896 EDS44453.1 GDIP01222332 JAJ01070.1 KQ423905 KOF71962.1 GDIQ01045111 JAN49626.1 NEDP02005475 OWF40323.1 QCYY01003913 ROT61933.1 EAA05194.3 GG666710 EEN42796.1 GFJQ02000885 JAW06085.1 KV921393 ORE16218.1 PJQL01000544 RCH94736.1 PJQL01000876 RCH92104.1 AMYB01000001 OAD07906.1 KZ303842 PHZ17944.1
JTDY01003664 KOB64137.1 KOB69190.1 JTDY01012558 KOB52256.1 GAIX01004416 JAA88144.1 AGBW02012542 OWR44851.1 PYGN01000705 PSN41939.1 KQ414619 KOC67725.1 KQ435903 KOX69083.1 KZ288280 PBC29640.1 AAZX01005171 KK853097 KDR11448.1 QOIP01000005 RLU22739.1 KQ971343 EFA04308.1 GL448228 EFN85094.1 KQ760974 OAD58574.1 NNAY01000869 OXU26135.1 GL771844 EFZ09450.1 ADTU01015804 ADTU01015805 ADTU01015806 KQ976737 KYM75907.1 GECZ01003136 JAS66633.1 GECU01008986 JAS98720.1 GL440027 EFN66391.1 LBMM01007854 KMQ89369.1 KQ976870 KYN07752.1 APGK01032712 KB740848 ENN78681.1 KB632411 ERL95236.1 BT127396 AEE62358.1 KQ981208 KYN44878.1 GEBQ01000889 JAT39088.1 KQ434868 KZC09171.1 GL888086 EGI67843.1 NEVH01010475 PNF32591.1 KQ982194 KYQ59065.1 KQ979763 KYN19234.1 GGMS01009546 MBY78749.1 GEDC01017130 JAS20168.1 GGMR01005374 MBY17993.1 ABLF02014276 GEZM01096214 JAV55049.1 GBBI01002521 JAC16191.1 GFDL01001418 JAV33627.1 ACPB03010315 GFDL01001413 JAV33632.1 GFDF01000829 JAV13255.1 DS235219 EEB13389.1 AJVK01031256 AJVK01031257 CH477343 EAT43050.1 ODYU01011528 SOQ57318.1 GEZM01096216 JAV55046.1 AXCN02001805 APCN01004514 ADMH02001629 ETN61696.1 AAAB01008816 GDIP01229894 GDIP01221186 GDIP01161345 GDIP01154999 JAJ02216.1 GDIP01175352 JAJ48050.1 GBHO01022343 GBHO01006490 GDHC01012304 JAG21261.1 JAG37114.1 JAQ06325.1 JXUM01120466 KQ566395 KXJ70160.1 GDIP01175351 JAJ48051.1 GDIQ01257320 JAJ94404.1 GDIP01245655 JAI77746.1 AXCM01003607 GL732523 EFX89900.1 GDIQ01076554 JAN18183.1 CVRI01000027 CRK92402.1 GEZM01004857 JAV96292.1 DS231896 EDS44453.1 GDIP01222332 JAJ01070.1 KQ423905 KOF71962.1 GDIQ01045111 JAN49626.1 NEDP02005475 OWF40323.1 QCYY01003913 ROT61933.1 EAA05194.3 GG666710 EEN42796.1 GFJQ02000885 JAW06085.1 KV921393 ORE16218.1 PJQL01000544 RCH94736.1 PJQL01000876 RCH92104.1 AMYB01000001 OAD07906.1 KZ303842 PHZ17944.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000007151
UP000245037
UP000005203
+ More
UP000053825 UP000053105 UP000242457 UP000002358 UP000027135 UP000279307 UP000007266 UP000008237 UP000215335 UP000005205 UP000078540 UP000000311 UP000036403 UP000078542 UP000019118 UP000030742 UP000078541 UP000076502 UP000007755 UP000235965 UP000075809 UP000078492 UP000007819 UP000192223 UP000015103 UP000009046 UP000092462 UP000008820 UP000075886 UP000075880 UP000076408 UP000075885 UP000075884 UP000075840 UP000069272 UP000000673 UP000075900 UP000076407 UP000075903 UP000075902 UP000095300 UP000069940 UP000249989 UP000075883 UP000000305 UP000183832 UP000075881 UP000075920 UP000002320 UP000053454 UP000242188 UP000283509 UP000007062 UP000001554 UP000075901 UP000242381 UP000252139 UP000077051 UP000242254
UP000053825 UP000053105 UP000242457 UP000002358 UP000027135 UP000279307 UP000007266 UP000008237 UP000215335 UP000005205 UP000078540 UP000000311 UP000036403 UP000078542 UP000019118 UP000030742 UP000078541 UP000076502 UP000007755 UP000235965 UP000075809 UP000078492 UP000007819 UP000192223 UP000015103 UP000009046 UP000092462 UP000008820 UP000075886 UP000075880 UP000076408 UP000075885 UP000075884 UP000075840 UP000069272 UP000000673 UP000075900 UP000076407 UP000075903 UP000075902 UP000095300 UP000069940 UP000249989 UP000075883 UP000000305 UP000183832 UP000075881 UP000075920 UP000002320 UP000053454 UP000242188 UP000283509 UP000007062 UP000001554 UP000075901 UP000242381 UP000252139 UP000077051 UP000242254
PRIDE
Interpro
SUPFAM
SSF140959
SSF140959
ProteinModelPortal
H9IV22
A0A3S2NNN1
A0A0L7KQ62
A0A0L7KLE8
A0A0L7K3A9
S4PBW5
+ More
A0A212ETS1 A0A2P8YCF1 A0A087ZSH3 A0A0L7RA36 A0A0M8ZT40 A0A2A3ED53 K7JCX9 A0A067R1I8 A0A3L8DSA7 D6WMV9 E2BGW2 A0A310SIC5 A0A232F677 E9JCK3 A0A158NHG6 A0A195AV25 A0A1B6GWA2 A0A1B6JHI6 E2AJJ6 A0A0J7KGJ8 A0A195D4E7 N6UDH8 U4UXZ2 J3JVX8 A0A195FWR3 A0A1B6MT62 A0A154PBE8 F4WDB6 A0A2J7QVI0 A0A151XFF3 A0A195E2X2 A0A2S2QLV1 A0A1B6D3B5 A0A2S2NLC4 J9JRN9 A0A1Y1K0N5 A0A1W4X9R6 A0A023F415 A0A1Q3G1F4 T1HF81 A0A1W4XK61 A0A1Q3G1F2 A0A1L8E3Z1 E0VJ33 A0A1B0FY58 Q179Y5 A0A1S4FAQ9 A0A2H1WW88 A0A1Y1K6K6 A0A182Q6V2 A0A182JLZ3 A0A182YIV0 A0A182PWE4 A0A182NV45 A0A182HYB5 A0A182FVE9 W5JC44 A0A182REL8 A0A1S4H781 A0A182WYS0 A0A182UMW5 A0A182TF14 A0A0P4YYC7 A0A0P5C6U4 A0A0A9YY67 A0A1I8PX39 A0A182H9D4 A0A0P5CHJ2 A0A0P5FBW7 A0A0P4XWI3 A0A182MEU3 E9FRT0 A0A0P6DK33 A0A1J1HXX0 A0A182JSR7 A0A182W9M6 A0A1Y1NEH3 B0WDE0 A0A0P4ZRJ3 A0A0L8G4J0 A0A0P6FP95 A0A210PV17 A0A3R7PWR9 Q7QHN7 C3ZXB6 A0A182SMQ4 A0A1Z5LFC9 A0A1X0RWD4 A0A367JXR2 A0A367JQ78 A0A168PLQ4 A0A2G4TA98
A0A212ETS1 A0A2P8YCF1 A0A087ZSH3 A0A0L7RA36 A0A0M8ZT40 A0A2A3ED53 K7JCX9 A0A067R1I8 A0A3L8DSA7 D6WMV9 E2BGW2 A0A310SIC5 A0A232F677 E9JCK3 A0A158NHG6 A0A195AV25 A0A1B6GWA2 A0A1B6JHI6 E2AJJ6 A0A0J7KGJ8 A0A195D4E7 N6UDH8 U4UXZ2 J3JVX8 A0A195FWR3 A0A1B6MT62 A0A154PBE8 F4WDB6 A0A2J7QVI0 A0A151XFF3 A0A195E2X2 A0A2S2QLV1 A0A1B6D3B5 A0A2S2NLC4 J9JRN9 A0A1Y1K0N5 A0A1W4X9R6 A0A023F415 A0A1Q3G1F4 T1HF81 A0A1W4XK61 A0A1Q3G1F2 A0A1L8E3Z1 E0VJ33 A0A1B0FY58 Q179Y5 A0A1S4FAQ9 A0A2H1WW88 A0A1Y1K6K6 A0A182Q6V2 A0A182JLZ3 A0A182YIV0 A0A182PWE4 A0A182NV45 A0A182HYB5 A0A182FVE9 W5JC44 A0A182REL8 A0A1S4H781 A0A182WYS0 A0A182UMW5 A0A182TF14 A0A0P4YYC7 A0A0P5C6U4 A0A0A9YY67 A0A1I8PX39 A0A182H9D4 A0A0P5CHJ2 A0A0P5FBW7 A0A0P4XWI3 A0A182MEU3 E9FRT0 A0A0P6DK33 A0A1J1HXX0 A0A182JSR7 A0A182W9M6 A0A1Y1NEH3 B0WDE0 A0A0P4ZRJ3 A0A0L8G4J0 A0A0P6FP95 A0A210PV17 A0A3R7PWR9 Q7QHN7 C3ZXB6 A0A182SMQ4 A0A1Z5LFC9 A0A1X0RWD4 A0A367JXR2 A0A367JQ78 A0A168PLQ4 A0A2G4TA98
PDB
3MBO
E-value=3.21412e-09,
Score=148
Ontologies
PATHWAY
GO
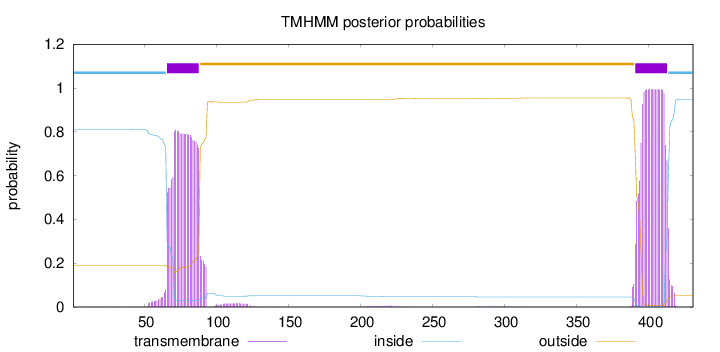
Topology
Length:
431
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
39.65253
Exp number, first 60 AAs:
0.21752
Total prob of N-in:
0.81080
inside
1 - 65
TMhelix
66 - 88
outside
89 - 390
TMhelix
391 - 413
inside
414 - 431
Population Genetic Test Statistics
Pi
18.643446
Theta
16.607939
Tajima's D
0.386153
CLR
0.359831
CSRT
0.483925803709815
Interpretation
Uncertain
