Gene
KWMTBOMO08105
Pre Gene Modal
BGIBMGA001103
Annotation
PREDICTED:_growth_arrest_and_DNA_damage-inducible_proteins-interacting_protein_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.531
Sequence
CDS
ATGAATTTGTGTGTTAGATTGAAATGCTTCCGTTATAACATTTCCAGCCATTTATACAGATATTCTACAACATCGGAAGCAATTGAACAAAATGAACAAGTAATTATTGACGACAGCGATGTAGCACAGGCGAAGGAAGAAGAAATTAATAAAAAGAGAAATATATCTCGTCTTAGTATGTCACATCATAACCTCGTTAACGGGAAACGACCTTACGACTATCCAATGAGTTTAGCTCATTTGACCGTGAAATACAACAGGAAAATGTATGGAAAATATGGAAGCGCTAGTGGAGTGAACCCTAGCTTATGCTGGCCAACTAGAGCAGACATCAGAGAGAAATTGGAATATGAATCCGAAGCATACCCTTTCACAATACAAGAGATGATGGAGACAACCAGGCAGAAACGTCTAGCTGAAGAGGAAAAGATACTGAAGAGAGATCAGGAAATTGTAGCCAAAATGGCAAAATTAGAGATGTGGAAAAAGGAACTTCGAAACAAAGTCGCCAAGAAAACTGCTGAAGCACAGGCTGCTAAGGACAAGAAGGAACGCCTTGTTGAGGAGGTGCGAAGACATTTTGGTTTCAAACTGGATTCTCGTGATGAAAGGTTCCAGGAGATGTTGGTCAAACGCGAGAAAGAACAAAAGAAACAGGAAAAACTAGCTAGAAAAGAAGCTAAGGAGAAGGTTATGATTGCCAAACTACAACAGAAGAATGCTGAAATCAGTGAAAACAAATAA
Protein
MNLCVRLKCFRYNISSHLYRYSTTSEAIEQNEQVIIDDSDVAQAKEEEINKKRNISRLSMSHHNLVNGKRPYDYPMSLAHLTVKYNRKMYGKYGSASGVNPSLCWPTRADIREKLEYESEAYPFTIQEMMETTRQKRLAEEEKILKRDQEIVAKMAKLEMWKKELRNKVAKKTAEAQAAKDKKERLVEEVRRHFGFKLDSRDERFQEMLVKREKEQKKQEKLARKEAKEKVMIAKLQQKNAEISENK
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A2H1WW83
A0A2A4JWT0
A0A194REF7
A0A194Q7R2
A0A212ETP6
S4PBA1
+ More
A0A437BPL4 A0A0L7KQ96 A0A1Y1KJ71 A0A2J7QI39 A0A1B6EWH5 A0A1B6HR31 A0A2P8XEE9 A0A336MJT9 A0A0A9WQI7 A0A1D2N747 A0A1B6MNJ5 A0A226EJU1 A0A1L8DL07 A0A1L8DKR8 A0A336LYX0 A0A1Q3F958 A0A1B0AR99 A0A1A9WTJ7 A0A1A9YL25 A0A2M3ZI99 A0A182Q1F8 A0A1Q3FDX4 W5JU54 A0A1A9ZFZ3 B0WHI4 A0A2M4BWL1 A0A2M4BWN1 A0A2M4BWC0 A0A182FBY9 A0A1B0FKN5 A0A1A9V304 A0A2M4BWA3 A0A1B0D544 A0A2M4API8 A0A0K8TRI4 A0A182N3B5 A0A069DY32 T1DIB1 A0A0L0CQK2 A0A084VHX7 A0A182JH78 A0A1B0CJY1 A0A182Y714 A0A182KFN3 A0A182GT36 A0A182GXP2 E2B1J8 A0A182UP41 A0A182WXG5 Q7QHT3 A0A182HY41 A0A182LH86 A0A182U940 N6TX77 A0A182PTY6 B4KY16 J3JUV1 A0A182RLI1 A0A182SEM2 A0A1I8N3P7 Q17B01 A0A182W1N3 A0A3L8DUW7 A0A182M0B9 A0A1S4F9T1 A0A1I8P6F7 B4IWX9 A0A158NC23 A0A195BG89
A0A437BPL4 A0A0L7KQ96 A0A1Y1KJ71 A0A2J7QI39 A0A1B6EWH5 A0A1B6HR31 A0A2P8XEE9 A0A336MJT9 A0A0A9WQI7 A0A1D2N747 A0A1B6MNJ5 A0A226EJU1 A0A1L8DL07 A0A1L8DKR8 A0A336LYX0 A0A1Q3F958 A0A1B0AR99 A0A1A9WTJ7 A0A1A9YL25 A0A2M3ZI99 A0A182Q1F8 A0A1Q3FDX4 W5JU54 A0A1A9ZFZ3 B0WHI4 A0A2M4BWL1 A0A2M4BWN1 A0A2M4BWC0 A0A182FBY9 A0A1B0FKN5 A0A1A9V304 A0A2M4BWA3 A0A1B0D544 A0A2M4API8 A0A0K8TRI4 A0A182N3B5 A0A069DY32 T1DIB1 A0A0L0CQK2 A0A084VHX7 A0A182JH78 A0A1B0CJY1 A0A182Y714 A0A182KFN3 A0A182GT36 A0A182GXP2 E2B1J8 A0A182UP41 A0A182WXG5 Q7QHT3 A0A182HY41 A0A182LH86 A0A182U940 N6TX77 A0A182PTY6 B4KY16 J3JUV1 A0A182RLI1 A0A182SEM2 A0A1I8N3P7 Q17B01 A0A182W1N3 A0A3L8DUW7 A0A182M0B9 A0A1S4F9T1 A0A1I8P6F7 B4IWX9 A0A158NC23 A0A195BG89
Pubmed
EMBL
ODYU01011528
SOQ57320.1
NWSH01000457
PCG76269.1
KQ460297
KPJ16218.1
+ More
KQ459439 KPJ01035.1 AGBW02012542 OWR44849.1 GAIX01006017 JAA86543.1 RSAL01000023 RVE52265.1 JTDY01007400 KOB65236.1 GEZM01081920 JAV61479.1 NEVH01013960 PNF28250.1 GECZ01027434 JAS42335.1 GECU01030567 JAS77139.1 PYGN01002518 PSN30352.1 UFQT01001464 SSX30672.1 GBHO01033928 GBRD01010004 GDHC01004778 JAG09676.1 JAG55820.1 JAQ13851.1 LJIJ01000174 ODN01052.1 GEBQ01002505 JAT37472.1 LNIX01000003 OXA57021.1 GFDF01007029 JAV07055.1 GFDF01007028 JAV07056.1 UFQT01000317 SSX23182.1 GFDL01010945 JAV24100.1 JXJN01002295 GGFM01007523 MBW28274.1 AXCN02001116 GFDL01009298 JAV25747.1 ADMH02000442 ETN66470.1 DS231936 EDS27762.1 GGFJ01008329 MBW57470.1 GGFJ01008328 MBW57469.1 GGFJ01008239 MBW57380.1 CCAG010023454 CCAG010023455 GGFJ01007867 MBW57008.1 AJVK01011711 GGFK01009362 MBW42683.1 GDAI01000624 JAI16979.1 GBGD01002610 JAC86279.1 GAMD01001976 JAA99614.1 JRES01000064 KNC34452.1 ATLV01013242 KE524847 KFB37571.1 AJWK01015525 JXUM01086155 KQ563548 KXJ73678.1 JXUM01095976 KQ564234 KXJ72551.1 GL444934 EFN60455.1 AAAB01008816 EAA05241.3 APCN01004490 APGK01047882 KB741092 KB632050 ENN73920.1 ERL88311.1 CH933809 EDW18718.1 BT127016 AEE61978.1 CH477328 EAT43413.1 QOIP01000004 RLU23983.1 AXCM01004566 CH916366 EDV97380.1 ADTU01011293 KQ976500 KYM83192.1
KQ459439 KPJ01035.1 AGBW02012542 OWR44849.1 GAIX01006017 JAA86543.1 RSAL01000023 RVE52265.1 JTDY01007400 KOB65236.1 GEZM01081920 JAV61479.1 NEVH01013960 PNF28250.1 GECZ01027434 JAS42335.1 GECU01030567 JAS77139.1 PYGN01002518 PSN30352.1 UFQT01001464 SSX30672.1 GBHO01033928 GBRD01010004 GDHC01004778 JAG09676.1 JAG55820.1 JAQ13851.1 LJIJ01000174 ODN01052.1 GEBQ01002505 JAT37472.1 LNIX01000003 OXA57021.1 GFDF01007029 JAV07055.1 GFDF01007028 JAV07056.1 UFQT01000317 SSX23182.1 GFDL01010945 JAV24100.1 JXJN01002295 GGFM01007523 MBW28274.1 AXCN02001116 GFDL01009298 JAV25747.1 ADMH02000442 ETN66470.1 DS231936 EDS27762.1 GGFJ01008329 MBW57470.1 GGFJ01008328 MBW57469.1 GGFJ01008239 MBW57380.1 CCAG010023454 CCAG010023455 GGFJ01007867 MBW57008.1 AJVK01011711 GGFK01009362 MBW42683.1 GDAI01000624 JAI16979.1 GBGD01002610 JAC86279.1 GAMD01001976 JAA99614.1 JRES01000064 KNC34452.1 ATLV01013242 KE524847 KFB37571.1 AJWK01015525 JXUM01086155 KQ563548 KXJ73678.1 JXUM01095976 KQ564234 KXJ72551.1 GL444934 EFN60455.1 AAAB01008816 EAA05241.3 APCN01004490 APGK01047882 KB741092 KB632050 ENN73920.1 ERL88311.1 CH933809 EDW18718.1 BT127016 AEE61978.1 CH477328 EAT43413.1 QOIP01000004 RLU23983.1 AXCM01004566 CH916366 EDV97380.1 ADTU01011293 KQ976500 KYM83192.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
UP000037510
+ More
UP000235965 UP000245037 UP000094527 UP000198287 UP000092460 UP000091820 UP000092443 UP000075886 UP000000673 UP000092445 UP000002320 UP000069272 UP000092444 UP000078200 UP000092462 UP000075884 UP000037069 UP000030765 UP000075880 UP000092461 UP000076408 UP000075881 UP000069940 UP000249989 UP000000311 UP000075903 UP000076407 UP000007062 UP000075840 UP000075882 UP000075902 UP000019118 UP000030742 UP000075885 UP000009192 UP000075900 UP000075901 UP000095301 UP000008820 UP000075920 UP000279307 UP000075883 UP000095300 UP000001070 UP000005205 UP000078540
UP000235965 UP000245037 UP000094527 UP000198287 UP000092460 UP000091820 UP000092443 UP000075886 UP000000673 UP000092445 UP000002320 UP000069272 UP000092444 UP000078200 UP000092462 UP000075884 UP000037069 UP000030765 UP000075880 UP000092461 UP000076408 UP000075881 UP000069940 UP000249989 UP000000311 UP000075903 UP000076407 UP000007062 UP000075840 UP000075882 UP000075902 UP000019118 UP000030742 UP000075885 UP000009192 UP000075900 UP000075901 UP000095301 UP000008820 UP000075920 UP000279307 UP000075883 UP000095300 UP000001070 UP000005205 UP000078540
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1WW83
A0A2A4JWT0
A0A194REF7
A0A194Q7R2
A0A212ETP6
S4PBA1
+ More
A0A437BPL4 A0A0L7KQ96 A0A1Y1KJ71 A0A2J7QI39 A0A1B6EWH5 A0A1B6HR31 A0A2P8XEE9 A0A336MJT9 A0A0A9WQI7 A0A1D2N747 A0A1B6MNJ5 A0A226EJU1 A0A1L8DL07 A0A1L8DKR8 A0A336LYX0 A0A1Q3F958 A0A1B0AR99 A0A1A9WTJ7 A0A1A9YL25 A0A2M3ZI99 A0A182Q1F8 A0A1Q3FDX4 W5JU54 A0A1A9ZFZ3 B0WHI4 A0A2M4BWL1 A0A2M4BWN1 A0A2M4BWC0 A0A182FBY9 A0A1B0FKN5 A0A1A9V304 A0A2M4BWA3 A0A1B0D544 A0A2M4API8 A0A0K8TRI4 A0A182N3B5 A0A069DY32 T1DIB1 A0A0L0CQK2 A0A084VHX7 A0A182JH78 A0A1B0CJY1 A0A182Y714 A0A182KFN3 A0A182GT36 A0A182GXP2 E2B1J8 A0A182UP41 A0A182WXG5 Q7QHT3 A0A182HY41 A0A182LH86 A0A182U940 N6TX77 A0A182PTY6 B4KY16 J3JUV1 A0A182RLI1 A0A182SEM2 A0A1I8N3P7 Q17B01 A0A182W1N3 A0A3L8DUW7 A0A182M0B9 A0A1S4F9T1 A0A1I8P6F7 B4IWX9 A0A158NC23 A0A195BG89
A0A437BPL4 A0A0L7KQ96 A0A1Y1KJ71 A0A2J7QI39 A0A1B6EWH5 A0A1B6HR31 A0A2P8XEE9 A0A336MJT9 A0A0A9WQI7 A0A1D2N747 A0A1B6MNJ5 A0A226EJU1 A0A1L8DL07 A0A1L8DKR8 A0A336LYX0 A0A1Q3F958 A0A1B0AR99 A0A1A9WTJ7 A0A1A9YL25 A0A2M3ZI99 A0A182Q1F8 A0A1Q3FDX4 W5JU54 A0A1A9ZFZ3 B0WHI4 A0A2M4BWL1 A0A2M4BWN1 A0A2M4BWC0 A0A182FBY9 A0A1B0FKN5 A0A1A9V304 A0A2M4BWA3 A0A1B0D544 A0A2M4API8 A0A0K8TRI4 A0A182N3B5 A0A069DY32 T1DIB1 A0A0L0CQK2 A0A084VHX7 A0A182JH78 A0A1B0CJY1 A0A182Y714 A0A182KFN3 A0A182GT36 A0A182GXP2 E2B1J8 A0A182UP41 A0A182WXG5 Q7QHT3 A0A182HY41 A0A182LH86 A0A182U940 N6TX77 A0A182PTY6 B4KY16 J3JUV1 A0A182RLI1 A0A182SEM2 A0A1I8N3P7 Q17B01 A0A182W1N3 A0A3L8DUW7 A0A182M0B9 A0A1S4F9T1 A0A1I8P6F7 B4IWX9 A0A158NC23 A0A195BG89
PDB
6NU3
E-value=1.29343e-11,
Score=166
Ontologies
GO
PANTHER
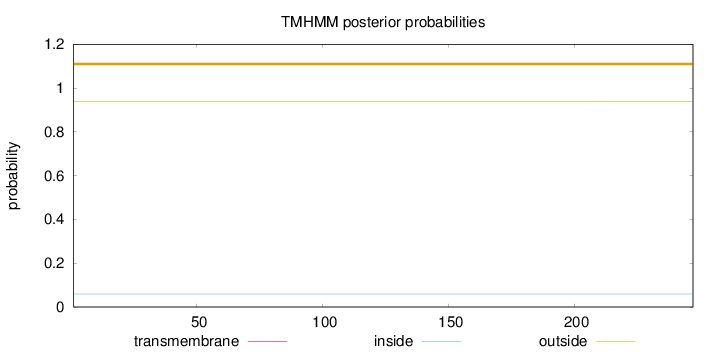
Topology
Length:
247
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00039
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05979
outside
1 - 247
Population Genetic Test Statistics
Pi
19.698365
Theta
22.926771
Tajima's D
-0.450568
CLR
0.685557
CSRT
0.251587420628969
Interpretation
Uncertain
