Gene
KWMTBOMO08103
Pre Gene Modal
BGIBMGA001056
Annotation
adipose_triglyceride_lipase_[Manduca_sexta]
Location in the cell
Nuclear Reliability : 2.57
Sequence
CDS
ATGAACTTATCGTTCGCTGGTTGTGGCTTCCTCGGGATTTACCATGTTGGTGTTGCAGTTTGTTTCAAGAAATATGCGCCTCATCTCCTTCTAGGGAAGATATCAGGAGCTTCTTTTGGAGCTTTGTCAGCCTGTTGCTTGCTCTGTGATTTGCCCATAGGTGAAATCACATCTGATGTTCTCAGAGTGGTCCGCGAAGCAAGGTCAGGGTCCCTCGGACCCTTTAGCCCTTCATTCAACATTCAAAATGTGCTCCTTGAGGGCATGGAGAAATATTTACCCGAAGACGCCCATAAGATAGTGAGCGGTAAACTACATATATCACTAACCAGAGTATACGATGGGAAGAATGTTATTGTATCGGAGTTCCCTACAAGAGAAGATTTATTACAGGCGGTGATGGCGTCGTGCTTTGTCCCAGTATTTTCTGGCCTGTTGCCCCCCCGATTCCACGGCATCAGGTATATGGATGGAGGGTTCAGTGATAACCTGCCAGTGTTAGATGAGAACACCATCACGGTCAGTCCCTTCTGCGGGGAGAGCGATATATGTCCTAGGGATTTGAGTTCACAGTTATTCCACGTAAACCTAGCGAACACAAGTATAGAGCTATCAAAACAAAACATGAATCGATTCGCTCGCATCCTTTTCCCTCCAAAACCCGAAGTCTTAAGCAACATGTGCAAGCAAGGCTTCGACGACGCTCTTCGATTCTTGCACAGAAACAACCTGATATCTTGCACGAGATGTCTCGCCGTGCAGTCGACCTTCCAGCTTCAAGACGTCTTGGATGACACAACGTACGATTATGATCCGGATTGTGAAGAGTGCAAGACCCATAGACAGGATGCATTAGTGGACGATCTACCGGACACGGTGATGACGATATTCCAAAATGCCATCGATTCGGCCAACAACGGCATCGTGAACTGGGTGATGAAGCAACGTGCCGTCCGATACCTGAACCTCCTGACGCTGCCGTACCGAGTGCCCATCGACATCATGTATGCTACATTCACCAAATTCGTATCATGCACGCCGAAGATGAGCAAGTCAGTCTGGAAGCTAAGCCTCAATCTCCTGCAGCAGCTCCACGGCTTCGTGTACACGTCGCACGACAGGTCCGAAGCCGCGGCCAGGATATACTACAAGCTGACCACCGAGGCGAAGAGGGAAACCAACGAAGGACACCTCTTGGAGAGGCGACGAGCGAGTGAGTTCCGTGTGACGTACGGTGACGTGAGAATAACCTATGAGGATGCGGACACCTTCGAACAAATACTCAACGTGACGTCACACCACGATGCACTGCTCGCATACTACTACCTCGACGGAGAGAATAAGATGAAGATGACCGAGATATACGACGTGACCGATGCCGACACGGACGCTGTGCAGTCGCCCACAGAGCGGGACCTCAACAAGCAGCTGGAGTTCGACAACGACTGGTCCGACGAACTCATCACCAGTAACGAGGAACTGGACATGGAAGCGATGGACGATGACGCCCTGGCCGACCGGAACATATTCTCGGACCCGGAGAGCGAGTGGAACGGGCGCCGATCCAACTCGGAGAGCGACGCCGAGCAACCGGAGGCCGACCGCCGCCCCCACGGGAGCGCCTACCTCGCCTACGCCGACTGA
Protein
MNLSFAGCGFLGIYHVGVAVCFKKYAPHLLLGKISGASFGALSACCLLCDLPIGEITSDVLRVVREARSGSLGPFSPSFNIQNVLLEGMEKYLPEDAHKIVSGKLHISLTRVYDGKNVIVSEFPTREDLLQAVMASCFVPVFSGLLPPRFHGIRYMDGGFSDNLPVLDENTITVSPFCGESDICPRDLSSQLFHVNLANTSIELSKQNMNRFARILFPPKPEVLSNMCKQGFDDALRFLHRNNLISCTRCLAVQSTFQLQDVLDDTTYDYDPDCEECKTHRQDALVDDLPDTVMTIFQNAIDSANNGIVNWVMKQRAVRYLNLLTLPYRVPIDIMYATFTKFVSCTPKMSKSVWKLSLNLLQQLHGFVYTSHDRSEAAARIYYKLTTEAKRETNEGHLLERRRASEFRVTYGDVRITYEDADTFEQILNVTSHHDALLAYYYLDGENKMKMTEIYDVTDADTDAVQSPTERDLNKQLEFDNDWSDELITSNEELDMEAMDDDALADRNIFSDPESEWNGRRSNSESDAEQPEADRRPHGSAYLAYAD
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
W0AME9
A0A291P0W9
F8UKL8
A0A194Q677
A0A212ETQ3
V9I9I8
+ More
V9I9G5 Q16NN1 D2A2L1 A0A084WIJ0 A0A2J7PGA8 A0A1Q3FX40 A0A1Q3G214 A0A1Q3FWT4 A0A1Q3FX08 A0A182JAF4 U5EQJ9 A0A2M4CP65 A0A2M4A2G2 A0A2M4A2N0 A0A151WSY5 T1HDH7 A0A2M4BHE2 A0A2M3ZG80 A0A2M4BHB4 A0A023F1Z7 A0A182Y9H7 A0A195AV08 A0A310SIB1 A0A195FMD6 A0A158NF51 A0A2M4BHB5 W5JW31 A0A1S4H7U8 A0A3F2YSQ6 A0A2M4CPJ2 E2AAK5 A0A026W335 A0A3L8DF05 A0A3Q9D0J8 A0A1I8Q9M1 A0A182ULQ6 A0A1Q3G212 A0A0M9A6G3 A0A182L557 A0A0R3P356 A0A3F2YSQ7 A0A0A1WK49 A0A0A1XK84 Q2M172 E0VV11 B3MAV8 A0A0A9W8S5 E1JI03 B4NM22 A0A0R1E2Q2 B4HHT5 A0A0Q5UJ04 Q9VUH7 B3ND61 B4PIH1 A0A0K8VVQ4 A0A0K8TSE0 A0A034W0G3 A0A034VZE4 A0A0M5J186 K7J107 V9IBB2 V9IBR1 B4JCS4 B4J1C3 A0A336K5T3 A0A067R959 A0A2M4A2W8 A0A0K8T3P4 A0A1B6L7K1 A0A182PTV1 A0A1B6C163 A0A087ZRM8 A0A1B6BZU4 A0A182FB72 N6UE62 A0A182TP74 A0A1B6CWF3 A0A1B6K160 A0A1B6GS74
V9I9G5 Q16NN1 D2A2L1 A0A084WIJ0 A0A2J7PGA8 A0A1Q3FX40 A0A1Q3G214 A0A1Q3FWT4 A0A1Q3FX08 A0A182JAF4 U5EQJ9 A0A2M4CP65 A0A2M4A2G2 A0A2M4A2N0 A0A151WSY5 T1HDH7 A0A2M4BHE2 A0A2M3ZG80 A0A2M4BHB4 A0A023F1Z7 A0A182Y9H7 A0A195AV08 A0A310SIB1 A0A195FMD6 A0A158NF51 A0A2M4BHB5 W5JW31 A0A1S4H7U8 A0A3F2YSQ6 A0A2M4CPJ2 E2AAK5 A0A026W335 A0A3L8DF05 A0A3Q9D0J8 A0A1I8Q9M1 A0A182ULQ6 A0A1Q3G212 A0A0M9A6G3 A0A182L557 A0A0R3P356 A0A3F2YSQ7 A0A0A1WK49 A0A0A1XK84 Q2M172 E0VV11 B3MAV8 A0A0A9W8S5 E1JI03 B4NM22 A0A0R1E2Q2 B4HHT5 A0A0Q5UJ04 Q9VUH7 B3ND61 B4PIH1 A0A0K8VVQ4 A0A0K8TSE0 A0A034W0G3 A0A034VZE4 A0A0M5J186 K7J107 V9IBB2 V9IBR1 B4JCS4 B4J1C3 A0A336K5T3 A0A067R959 A0A2M4A2W8 A0A0K8T3P4 A0A1B6L7K1 A0A182PTV1 A0A1B6C163 A0A087ZRM8 A0A1B6BZU4 A0A182FB72 N6UE62 A0A182TP74 A0A1B6CWF3 A0A1B6K160 A0A1B6GS74
Pubmed
28986331
26354079
22118469
17510324
18362917
19820115
+ More
24438588 25474469 25244985 21347285 20920257 23761445 12364791 20798317 24508170 30249741 29956367 20966253 15632085 17994087 25830018 20566863 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356 26369729 25348373 20075255 24845553 23537049
24438588 25474469 25244985 21347285 20920257 23761445 12364791 20798317 24508170 30249741 29956367 20966253 15632085 17994087 25830018 20566863 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356 26369729 25348373 20075255 24845553 23537049
EMBL
KF724681
AHE57674.1
MF687631
ATJ44557.1
JF809663
AEJ33048.1
+ More
KQ459439 KPJ01037.1 AGBW02012542 OWR44847.1 JR037406 AEY57755.1 JR037404 AEY57753.1 CH477816 EAT35954.1 KQ971338 EFA02006.1 ATLV01023934 KE525347 KFB50034.1 NEVH01025635 PNF15364.1 GFDL01002999 JAV32046.1 GFDL01001188 JAV33857.1 GFDL01002975 JAV32070.1 GFDL01002967 JAV32078.1 GANO01003236 JAB56635.1 GGFL01002948 MBW67126.1 GGFK01001589 MBW34910.1 GGFK01001587 MBW34908.1 KQ982769 KYQ50948.1 ACPB03000725 GGFJ01003303 MBW52444.1 GGFM01006768 MBW27519.1 GGFJ01003305 MBW52446.1 GBBI01003125 JAC15587.1 KQ976736 KYM76011.1 KQ759874 OAD62361.1 KQ981490 KYN41397.1 ADTU01013745 ADTU01013746 ADTU01013747 GGFJ01003304 MBW52445.1 ADMH02000120 ETN67723.1 AAAB01008986 APCN01002426 GGFL01003072 MBW67250.1 GL438128 EFN69552.1 KK107519 EZA49449.1 QOIP01000009 RLU18498.1 MH238459 AZP54623.1 GFDL01001205 JAV33840.1 KQ435740 KOX76829.1 CH379069 KRT07548.1 GBXI01015236 JAC99055.1 GBXI01002518 JAD11774.1 EAL30703.1 DS235800 EEB17217.1 CH902618 EDV39192.2 GBHO01039430 GDHC01019151 JAG04174.1 JAP99477.1 AE014296 ACZ94716.1 CH964278 EDW85390.1 CM000159 KRK01906.1 CH480815 EDW41500.1 CH954178 KQS43974.1 AY051668 AAF49704.1 AAK93092.1 EDV51783.1 EDW94528.1 GDHF01023447 GDHF01012214 GDHF01009360 JAI28867.1 JAI40100.1 JAI42954.1 GDAI01000324 JAI17279.1 GAKP01011730 GAKP01011729 JAC47223.1 GAKP01011732 JAC47220.1 CP012523 ALC38336.1 JR037407 AEY57756.1 JR037405 AEY57754.1 CH916368 EDW03163.1 CH916366 EDV97992.1 UFQS01000092 UFQT01000092 SSW99212.1 SSX19592.1 KK852614 KDR20213.1 GGFK01001739 MBW35060.1 GBRD01005825 JAG59996.1 GEBQ01020342 JAT19635.1 GEDC01030308 JAS06990.1 GEDC01030517 GEDC01016531 JAS06781.1 JAS20767.1 APGK01026024 APGK01026025 APGK01026026 APGK01026027 APGK01026028 KB740612 ENN80000.1 GEDC01019765 JAS17533.1 GECU01002535 JAT05172.1 GECZ01004491 JAS65278.1
KQ459439 KPJ01037.1 AGBW02012542 OWR44847.1 JR037406 AEY57755.1 JR037404 AEY57753.1 CH477816 EAT35954.1 KQ971338 EFA02006.1 ATLV01023934 KE525347 KFB50034.1 NEVH01025635 PNF15364.1 GFDL01002999 JAV32046.1 GFDL01001188 JAV33857.1 GFDL01002975 JAV32070.1 GFDL01002967 JAV32078.1 GANO01003236 JAB56635.1 GGFL01002948 MBW67126.1 GGFK01001589 MBW34910.1 GGFK01001587 MBW34908.1 KQ982769 KYQ50948.1 ACPB03000725 GGFJ01003303 MBW52444.1 GGFM01006768 MBW27519.1 GGFJ01003305 MBW52446.1 GBBI01003125 JAC15587.1 KQ976736 KYM76011.1 KQ759874 OAD62361.1 KQ981490 KYN41397.1 ADTU01013745 ADTU01013746 ADTU01013747 GGFJ01003304 MBW52445.1 ADMH02000120 ETN67723.1 AAAB01008986 APCN01002426 GGFL01003072 MBW67250.1 GL438128 EFN69552.1 KK107519 EZA49449.1 QOIP01000009 RLU18498.1 MH238459 AZP54623.1 GFDL01001205 JAV33840.1 KQ435740 KOX76829.1 CH379069 KRT07548.1 GBXI01015236 JAC99055.1 GBXI01002518 JAD11774.1 EAL30703.1 DS235800 EEB17217.1 CH902618 EDV39192.2 GBHO01039430 GDHC01019151 JAG04174.1 JAP99477.1 AE014296 ACZ94716.1 CH964278 EDW85390.1 CM000159 KRK01906.1 CH480815 EDW41500.1 CH954178 KQS43974.1 AY051668 AAF49704.1 AAK93092.1 EDV51783.1 EDW94528.1 GDHF01023447 GDHF01012214 GDHF01009360 JAI28867.1 JAI40100.1 JAI42954.1 GDAI01000324 JAI17279.1 GAKP01011730 GAKP01011729 JAC47223.1 GAKP01011732 JAC47220.1 CP012523 ALC38336.1 JR037407 AEY57756.1 JR037405 AEY57754.1 CH916368 EDW03163.1 CH916366 EDV97992.1 UFQS01000092 UFQT01000092 SSW99212.1 SSX19592.1 KK852614 KDR20213.1 GGFK01001739 MBW35060.1 GBRD01005825 JAG59996.1 GEBQ01020342 JAT19635.1 GEDC01030308 JAS06990.1 GEDC01030517 GEDC01016531 JAS06781.1 JAS20767.1 APGK01026024 APGK01026025 APGK01026026 APGK01026027 APGK01026028 KB740612 ENN80000.1 GEDC01019765 JAS17533.1 GECU01002535 JAT05172.1 GECZ01004491 JAS65278.1
Proteomes
UP000053268
UP000007151
UP000008820
UP000007266
UP000030765
UP000235965
+ More
UP000075880 UP000075809 UP000015103 UP000076408 UP000078540 UP000078541 UP000005205 UP000000673 UP000075840 UP000000311 UP000053097 UP000279307 UP000095300 UP000075903 UP000053105 UP000075882 UP000001819 UP000009046 UP000007801 UP000000803 UP000007798 UP000002282 UP000001292 UP000008711 UP000092553 UP000002358 UP000001070 UP000027135 UP000075885 UP000005203 UP000069272 UP000019118 UP000075902
UP000075880 UP000075809 UP000015103 UP000076408 UP000078540 UP000078541 UP000005205 UP000000673 UP000075840 UP000000311 UP000053097 UP000279307 UP000095300 UP000075903 UP000053105 UP000075882 UP000001819 UP000009046 UP000007801 UP000000803 UP000007798 UP000002282 UP000001292 UP000008711 UP000092553 UP000002358 UP000001070 UP000027135 UP000075885 UP000005203 UP000069272 UP000019118 UP000075902
Interpro
SUPFAM
SSF52151
SSF52151
ProteinModelPortal
W0AME9
A0A291P0W9
F8UKL8
A0A194Q677
A0A212ETQ3
V9I9I8
+ More
V9I9G5 Q16NN1 D2A2L1 A0A084WIJ0 A0A2J7PGA8 A0A1Q3FX40 A0A1Q3G214 A0A1Q3FWT4 A0A1Q3FX08 A0A182JAF4 U5EQJ9 A0A2M4CP65 A0A2M4A2G2 A0A2M4A2N0 A0A151WSY5 T1HDH7 A0A2M4BHE2 A0A2M3ZG80 A0A2M4BHB4 A0A023F1Z7 A0A182Y9H7 A0A195AV08 A0A310SIB1 A0A195FMD6 A0A158NF51 A0A2M4BHB5 W5JW31 A0A1S4H7U8 A0A3F2YSQ6 A0A2M4CPJ2 E2AAK5 A0A026W335 A0A3L8DF05 A0A3Q9D0J8 A0A1I8Q9M1 A0A182ULQ6 A0A1Q3G212 A0A0M9A6G3 A0A182L557 A0A0R3P356 A0A3F2YSQ7 A0A0A1WK49 A0A0A1XK84 Q2M172 E0VV11 B3MAV8 A0A0A9W8S5 E1JI03 B4NM22 A0A0R1E2Q2 B4HHT5 A0A0Q5UJ04 Q9VUH7 B3ND61 B4PIH1 A0A0K8VVQ4 A0A0K8TSE0 A0A034W0G3 A0A034VZE4 A0A0M5J186 K7J107 V9IBB2 V9IBR1 B4JCS4 B4J1C3 A0A336K5T3 A0A067R959 A0A2M4A2W8 A0A0K8T3P4 A0A1B6L7K1 A0A182PTV1 A0A1B6C163 A0A087ZRM8 A0A1B6BZU4 A0A182FB72 N6UE62 A0A182TP74 A0A1B6CWF3 A0A1B6K160 A0A1B6GS74
V9I9G5 Q16NN1 D2A2L1 A0A084WIJ0 A0A2J7PGA8 A0A1Q3FX40 A0A1Q3G214 A0A1Q3FWT4 A0A1Q3FX08 A0A182JAF4 U5EQJ9 A0A2M4CP65 A0A2M4A2G2 A0A2M4A2N0 A0A151WSY5 T1HDH7 A0A2M4BHE2 A0A2M3ZG80 A0A2M4BHB4 A0A023F1Z7 A0A182Y9H7 A0A195AV08 A0A310SIB1 A0A195FMD6 A0A158NF51 A0A2M4BHB5 W5JW31 A0A1S4H7U8 A0A3F2YSQ6 A0A2M4CPJ2 E2AAK5 A0A026W335 A0A3L8DF05 A0A3Q9D0J8 A0A1I8Q9M1 A0A182ULQ6 A0A1Q3G212 A0A0M9A6G3 A0A182L557 A0A0R3P356 A0A3F2YSQ7 A0A0A1WK49 A0A0A1XK84 Q2M172 E0VV11 B3MAV8 A0A0A9W8S5 E1JI03 B4NM22 A0A0R1E2Q2 B4HHT5 A0A0Q5UJ04 Q9VUH7 B3ND61 B4PIH1 A0A0K8VVQ4 A0A0K8TSE0 A0A034W0G3 A0A034VZE4 A0A0M5J186 K7J107 V9IBB2 V9IBR1 B4JCS4 B4J1C3 A0A336K5T3 A0A067R959 A0A2M4A2W8 A0A0K8T3P4 A0A1B6L7K1 A0A182PTV1 A0A1B6C163 A0A087ZRM8 A0A1B6BZU4 A0A182FB72 N6UE62 A0A182TP74 A0A1B6CWF3 A0A1B6K160 A0A1B6GS74
Ontologies
GO
PANTHER
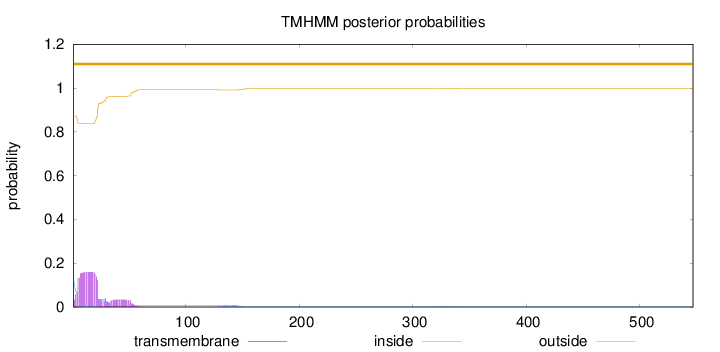
Topology
Length:
547
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.06726
Exp number, first 60 AAs:
3.87179
Total prob of N-in:
0.12776
outside
1 - 547
Population Genetic Test Statistics
Pi
19.031629
Theta
18.140859
Tajima's D
-1.033441
CLR
1.990373
CSRT
0.128893555322234
Interpretation
Uncertain
