Gene
KWMTBOMO08099
Pre Gene Modal
BGIBMGA001054
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_46_isoform_X1_[Bombyx_mori]
Full name
Ubiquitin carboxyl-terminal hydrolase 46
+ More
Ubiquitin carboxyl-terminal hydrolase 12
Ubiquitin carboxyl-terminal hydrolase 12A
Ubiquitin carboxyl-terminal hydrolase 12-A
Ubiquitin carboxyl-terminal hydrolase 12-B
Ubiquitin carboxyl-terminal hydrolase 4
Ubiquitin carboxyl-terminal hydrolase 3
Palmitoyltransferase
Probable ubiquitin carboxyl-terminal hydrolase creB
Ubiquitin carboxyl-terminal hydrolase 12
Ubiquitin carboxyl-terminal hydrolase 12A
Ubiquitin carboxyl-terminal hydrolase 12-A
Ubiquitin carboxyl-terminal hydrolase 12-B
Ubiquitin carboxyl-terminal hydrolase 4
Ubiquitin carboxyl-terminal hydrolase 3
Palmitoyltransferase
Probable ubiquitin carboxyl-terminal hydrolase creB
Alternative Name
Deubiquitinating enzyme 46
Ubiquitin thioesterase 46
Ubiquitin-specific-processing protease 46
Deubiquitinating enzyme 12
Ubiquitin thioesterase 12
Ubiquitin-hydrolyzing enzyme 1
Ubiquitin-specific-processing protease 12
Deubiquitinating enzyme 12A
Ubiquitin thioesterase 12A
Ubiquitin-specific-processing protease 12A
Deubiquitinating enzyme 12-A
Ubiquitin thioesterase 12-A
Ubiquitin-specific-processing protease 12-A
Deubiquitinating enzyme 12-B
Ubiquitin thioesterase 12-B
Ubiquitin-specific-processing protease 12-B
Deubiquitinating enzyme
Ubiquitin thioesterase
Ubiquitin-specific-processing protease
Deubiquitinating enzyme 4
Ubiquitin thioesterase 4
Ubiquitin-specific-processing protease 4
Deubiquitinating enzyme 3
Ubiquitin thioesterase 3
Ubiquitin-specific-processing protease 3
Carbon catabolite repression protein B
Deubiquitinating enzyme creB
Ubiquitin thioesterase creB
Ubiquitin-hydrolyzing enzyme creB
Ubiquitin-specific-processing protease creB
Ubiquitin thioesterase 46
Ubiquitin-specific-processing protease 46
Deubiquitinating enzyme 12
Ubiquitin thioesterase 12
Ubiquitin-hydrolyzing enzyme 1
Ubiquitin-specific-processing protease 12
Deubiquitinating enzyme 12A
Ubiquitin thioesterase 12A
Ubiquitin-specific-processing protease 12A
Deubiquitinating enzyme 12-A
Ubiquitin thioesterase 12-A
Ubiquitin-specific-processing protease 12-A
Deubiquitinating enzyme 12-B
Ubiquitin thioesterase 12-B
Ubiquitin-specific-processing protease 12-B
Deubiquitinating enzyme
Ubiquitin thioesterase
Ubiquitin-specific-processing protease
Deubiquitinating enzyme 4
Ubiquitin thioesterase 4
Ubiquitin-specific-processing protease 4
Deubiquitinating enzyme 3
Ubiquitin thioesterase 3
Ubiquitin-specific-processing protease 3
Carbon catabolite repression protein B
Deubiquitinating enzyme creB
Ubiquitin thioesterase creB
Ubiquitin-hydrolyzing enzyme creB
Ubiquitin-specific-processing protease creB
Location in the cell
Nuclear Reliability : 3.525
Sequence
CDS
ATGTCAATATGTCAGGATTTTGGCCTTAATTTCCAGCATAGAATGGGCGCAAATATATCACAGTTGGAAAGAGATATCGGGTCAGAGCAGTTCCCACCTAACGAACATTATTTCGGATTAGTCAATTTTGGCAACACTTGTTACAGCAACTCAGTGTTGCAAGCTTTATATTTTTGTCGCCCATTCAGAGAGAAAGTATTAGAGTACAAAGCTAAGAATAAAAGGACCAAGGAAACTCTATTAACATGCCTGGCAGATTTATTTTATAGCATAGCTACTCAAAAGAAAAAAGTTGGATCAATTGCACCTAAAAAATTTATTGCTAGACTTAGGAAGGAAAAGGAGGAATTTGACAATTACATGCAACAAGATGCCCATGAATTCCTGAACTTCCTAATAAATCATATTAATGAAATTATTTTAGCGGAACGTAATCAAAGCACATTAAAAGTTCAAAAGAACAACTCCGGTGAGAATGTGACGTGTAATGGTTCAGTGCCTCACAACACTGAACCGACTTGGGTGCACGAAATATTCCAAGGAACATTAACAAGCGAGACGAGGTGTCTCAACTGTGAGACGGTCAGCAGTAAAGATGAGCATTTCTTTGATTTACAGGTGGACGTGGGTCAAAACACGAGCATCACGCACTGCCTCAAGTGCTTCAGCGACACGGAGACGCTTTGCAACGACAACAAGTTCAAGTGTGACAACTGCAGCAGCTACCAGGAGGCGCAGAAGCGGATGCGCGTGAAGAAGCTTCCCCTCATCCTCGCCCTGCACCTCAAGAGGTTCAAATACATGGAACAGTTTAACCGGCACATCAAGGTGTCGCATCGCGTTGTGTTCCCATTAGAACTGAGATTATTTAATACGTCTGACGACGCTGTGAATCCGGATCGTCTGTACGACCTGGTGGCCGTGGTGGTGCACTGCGGCTCCGGCCCGAACCGGGGCCACTACATCAGCATCGTCAAGAGTCACGGCTTCTGGCTGCTGTTCGACGATGACATGGTTGATAAAATAGATGCATCAGCTATAGAAGACTTTTACGGCCTAACTTCAGACATACAGAAATCGTCCGAAACAGGATACATACTATTTTATCAGTCTAGGGACGCTAGTTGTTAA
Protein
MSICQDFGLNFQHRMGANISQLERDIGSEQFPPNEHYFGLVNFGNTCYSNSVLQALYFCRPFREKVLEYKAKNKRTKETLLTCLADLFYSIATQKKKVGSIAPKKFIARLRKEKEEFDNYMQQDAHEFLNFLINHINEIILAERNQSTLKVQKNNSGENVTCNGSVPHNTEPTWVHEIFQGTLTSETRCLNCETVSSKDEHFFDLQVDVGQNTSITHCLKCFSDTETLCNDNKFKCDNCSSYQEAQKRMRVKKLPLILALHLKRFKYMEQFNRHIKVSHRVVFPLELRLFNTSDDAVNPDRLYDLVAVVVHCGSGPNRGHYISIVKSHGFWLLFDDDMVDKIDASAIEDFYGLTSDIQKSSETGYILFYQSRDASC
Summary
Description
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action (By similarity). Has almost no deubiquitinating activity by itself and requires the interaction with wdr48 to have a high activity (By similarity).
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action. May act by mediating the deubiquitination of GAD1/GAD67 (By similarity). Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity. Not involved in deubiquitination of monoubiquitinated FANCD2 (By similarity).
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action. May act by mediating the deubiquitination of GAD1/GAD67 (PubMed:19465912). Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity. Not involved in deubiquitination of monoubiquitinated FANCD2 (By similarity).
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action. May act by mediating the deubiquitination of GAD1/GAD67 (By similarity). Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity (PubMed:19075014, PubMed:26388029). Not involved in deubiquitination of monoubiquitinated FANCD2 (PubMed:19075014).
Deubiquitinating enzyme. Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity. Not involved in deubiquitination of monoubiquitinated FANCD2.
Deubiquitinating enzyme. Has almost no deubiquitinating activity by itself and requires the interaction with WDR20 and WDR48 to have a high activity (PubMed:19075014, PubMed:27373336). Not involved in deubiquitination of monoubiquitinated FANCD2 (PubMed:19075014). In complex with WDR48, acts as a potential tumor suppressor by positively regulating PHLPP1 stability (PubMed:24145035).
Deubiquitinating enzyme. Has almost no deubiquitinating activity by itself and requires the interaction with wdr48 to have a high activity.
Regulates the abundance of the glr-1 glutamate receptor in the ventral nerve cord by promoting its deubiquitination and preventing its degradation in the lysosome. Contributes to the regulation of embryonic polarity.
Recognizes and hydrolyzes the peptide bond at the C-terminal Gly of ubiquitin. Involved in the processing of poly-ubiquitin precursors as well as that of ubiquitinated proteins. Required for the correct development of pollen.
Ubiquitin thioesterase component of the regulatory network controlling carbon source utilization through ubiquitination and deubiquitination involving creA, creB, creC, creD and acrB. Deubiquitinates the creA catabolic repressor and the quinate permease qutD. Plays also a role in response to carbon starvation and the control of extracellular proteases activity (By similarity).
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action. May act by mediating the deubiquitination of GAD1/GAD67 (By similarity). Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity. Not involved in deubiquitination of monoubiquitinated FANCD2 (By similarity).
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action. May act by mediating the deubiquitination of GAD1/GAD67 (PubMed:19465912). Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity. Not involved in deubiquitination of monoubiquitinated FANCD2 (By similarity).
Deubiquitinating enzyme that plays a role in behavior, possibly by regulating GABA action. May act by mediating the deubiquitination of GAD1/GAD67 (By similarity). Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity (PubMed:19075014, PubMed:26388029). Not involved in deubiquitination of monoubiquitinated FANCD2 (PubMed:19075014).
Deubiquitinating enzyme. Has almost no deubiquitinating activity by itself and requires the interaction with WDR48 to have a high activity. Not involved in deubiquitination of monoubiquitinated FANCD2.
Deubiquitinating enzyme. Has almost no deubiquitinating activity by itself and requires the interaction with WDR20 and WDR48 to have a high activity (PubMed:19075014, PubMed:27373336). Not involved in deubiquitination of monoubiquitinated FANCD2 (PubMed:19075014). In complex with WDR48, acts as a potential tumor suppressor by positively regulating PHLPP1 stability (PubMed:24145035).
Deubiquitinating enzyme. Has almost no deubiquitinating activity by itself and requires the interaction with wdr48 to have a high activity.
Regulates the abundance of the glr-1 glutamate receptor in the ventral nerve cord by promoting its deubiquitination and preventing its degradation in the lysosome. Contributes to the regulation of embryonic polarity.
Recognizes and hydrolyzes the peptide bond at the C-terminal Gly of ubiquitin. Involved in the processing of poly-ubiquitin precursors as well as that of ubiquitinated proteins. Required for the correct development of pollen.
Ubiquitin thioesterase component of the regulatory network controlling carbon source utilization through ubiquitination and deubiquitination involving creA, creB, creC, creD and acrB. Deubiquitinates the creA catabolic repressor and the quinate permease qutD. Plays also a role in response to carbon starvation and the control of extracellular proteases activity (By similarity).
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Subunit
Interacts with WDR48.
Interacts with WDR48. Interacts with WDR20.
Interacts with WDR48 (PubMed:19075014, PubMed:26388029). Interacts with WDR20 (PubMed:20147737).
Interacts with WDR48. Interacts with WDR20. Component of the USP12/WDR20/WDR48 deubiquitinating complex.
Interacts with WDR48 (PubMed:19075014, PubMed:27650958, PubMed:27373336). Interacts with WDR20 (PubMed:20147737, PubMed:27373336). Component of the USP12/WDR20/WDR48 deubiquitinating complex (PubMed:20147737, PubMed:27373336). Interacts with PHLPP1 (PubMed:24145035).
Interacts with wdr-20 and wdr-48; the catalytic activity of usp-46 is increased in the presence of both wdr-20 and wdr-48. Interacts with glr-1; the interaction results in deubiquitination of glr-1.
Interacts with creA, creC and qutD.
Interacts with WDR48. Interacts with WDR20.
Interacts with WDR48 (PubMed:19075014, PubMed:26388029). Interacts with WDR20 (PubMed:20147737).
Interacts with WDR48. Interacts with WDR20. Component of the USP12/WDR20/WDR48 deubiquitinating complex.
Interacts with WDR48 (PubMed:19075014, PubMed:27650958, PubMed:27373336). Interacts with WDR20 (PubMed:20147737, PubMed:27373336). Component of the USP12/WDR20/WDR48 deubiquitinating complex (PubMed:20147737, PubMed:27373336). Interacts with PHLPP1 (PubMed:24145035).
Interacts with wdr-20 and wdr-48; the catalytic activity of usp-46 is increased in the presence of both wdr-20 and wdr-48. Interacts with glr-1; the interaction results in deubiquitination of glr-1.
Interacts with creA, creC and qutD.
Miscellaneous
Knockdown of USP12 increases Akt activation.
Similarity
Belongs to the peptidase C19 family. USP12/USP46 subfamily.
Belongs to the peptidase C19 family.
Belongs to the DHHC palmitoyltransferase family.
Belongs to the peptidase C19 family.
Belongs to the DHHC palmitoyltransferase family.
Keywords
Behavior
Complete proteome
Hydrolase
Metal-binding
Protease
Reference proteome
Thiol protease
Ubl conjugation pathway
Zinc
Alternative splicing
3D-structure
Polymorphism
Cytoplasm
Lipoprotein
Myristate
Nucleus
Coiled coil
Feature
chain Ubiquitin carboxyl-terminal hydrolase 46
splice variant In isoform 2.
sequence variant In dbSNP:rs17475800.
splice variant In isoform 2.
sequence variant In dbSNP:rs17475800.
Uniprot
A5WWB0
F1M625
Q5RBQ4
P62069
P62068
Q9D9M2
+ More
A5D9H7 O75317 A4FUN7 Q52KZ6 Q5M981 C0HB46 P34547 Q8LAM0 O24454 A0A2I1E4B4 A0A372R1S4 A0A2H5RZT2 A0A2N0S567 A0A022VRL4 F4P9S8 A0A2I1CM32 A0A166ATA3 B0CNB1 A0A067NWK7 K5Y6W7 A0A409VEF5 A0A0C3P1N6 C8VD90 A0A1U7LQB1 A0A166S0Z9 A0A137QS67 A0A1C7NMP4 A0A2N1P1T7 A0A0A1PD09 A0A1Y3MMV9 A0A2Z6RTU0 A0A0C7CJW2 A0A316V7D4 A0A0A1NTD1 A0A0A1PF16 A0A367K8G3 A0A0C7BF14 A0A367J515 A0A286UST7 I1CGV4 S2JRR9 A0A409W7B0 A0A1X2ICC7 A0A0C7CLL8 A0A077WYX3 A0A1X0RTB5 Q6CB96 A0A1B9GHD3 B8P5U4 A0A0B7NMT2 A0A1X2IVZ3 A0A068SC62 U5HCE2 A0A1X2H9H2 A0A1E1K971 A0A0L0VFN5 A0A0D7AJH2 A0A0C3S8P8 A0A2I1FTX9 A0A0B7FEQ6 A0A1X0QUH1 X8JVI7 A0A0L6UN82 A0A077RB21 A0A433PNY3 X0BKP4 A0A0A2VHI9 A0A165NBC2 E9CXL2 W9XD26 A0A2Z6RV19 A0A395J5Q6 F0U5R9 C6H2S0 A0A3M7DFA1 A0A317SD19 C4JKS7 A0A1F8A596 A0A1Y2M0T9 A0A165JBF0 A0A067QHM1 A0A3M7G508 A0A1Y1W4E6 A0A395J8G9 Q0CT11 A0A2P2HNM3 E4ZVE8 W9Y652 A0A1C7LMM6 A0A0F0I917 A0A3N4IZ82 Q2UUG8 A0A2H1FJX3 I8TFM0
A5D9H7 O75317 A4FUN7 Q52KZ6 Q5M981 C0HB46 P34547 Q8LAM0 O24454 A0A2I1E4B4 A0A372R1S4 A0A2H5RZT2 A0A2N0S567 A0A022VRL4 F4P9S8 A0A2I1CM32 A0A166ATA3 B0CNB1 A0A067NWK7 K5Y6W7 A0A409VEF5 A0A0C3P1N6 C8VD90 A0A1U7LQB1 A0A166S0Z9 A0A137QS67 A0A1C7NMP4 A0A2N1P1T7 A0A0A1PD09 A0A1Y3MMV9 A0A2Z6RTU0 A0A0C7CJW2 A0A316V7D4 A0A0A1NTD1 A0A0A1PF16 A0A367K8G3 A0A0C7BF14 A0A367J515 A0A286UST7 I1CGV4 S2JRR9 A0A409W7B0 A0A1X2ICC7 A0A0C7CLL8 A0A077WYX3 A0A1X0RTB5 Q6CB96 A0A1B9GHD3 B8P5U4 A0A0B7NMT2 A0A1X2IVZ3 A0A068SC62 U5HCE2 A0A1X2H9H2 A0A1E1K971 A0A0L0VFN5 A0A0D7AJH2 A0A0C3S8P8 A0A2I1FTX9 A0A0B7FEQ6 A0A1X0QUH1 X8JVI7 A0A0L6UN82 A0A077RB21 A0A433PNY3 X0BKP4 A0A0A2VHI9 A0A165NBC2 E9CXL2 W9XD26 A0A2Z6RV19 A0A395J5Q6 F0U5R9 C6H2S0 A0A3M7DFA1 A0A317SD19 C4JKS7 A0A1F8A596 A0A1Y2M0T9 A0A165JBF0 A0A067QHM1 A0A3M7G508 A0A1Y1W4E6 A0A395J8G9 Q0CT11 A0A2P2HNM3 E4ZVE8 W9Y652 A0A1C7LMM6 A0A0F0I917 A0A3N4IZ82 Q2UUG8 A0A2H1FJX3 I8TFM0
EC Number
3.4.19.12
2.3.1.225
2.3.1.225
Pubmed
23594743
22043315
15057822
16141072
15489334
19465912
+ More
14702039 14715245 19075014 20147737 26388029 27650958 12391724 16305752 9615226 15057823 24145035 27373336 20433749 7906398 9851916 21273419 23209443 24356955 9268021 10617197 27862469 14593172 10898935 11115897 12912986 17905865 10617198 24277808 30271968 29355972 26659563 18322534 24958869 23045686 30283667 16372000 19146970 29771364 29674435 19578406 27956601 15229592 19193860 26076695 25683379 25474575 25359908 25062916 30485448 20516208 29764372 19717792 27664179 26693682 21326234 30420746 16372010 22933657
14702039 14715245 19075014 20147737 26388029 27650958 12391724 16305752 9615226 15057823 24145035 27373336 20433749 7906398 9851916 21273419 23209443 24356955 9268021 10617197 27862469 14593172 10898935 11115897 12912986 17905865 10617198 24277808 30271968 29355972 26659563 18322534 24958869 23045686 30283667 16372000 19146970 29771364 29674435 19578406 27956601 15229592 19193860 26076695 25683379 25474575 25359908 25062916 30485448 20516208 29764372 19717792 27664179 26693682 21326234 30420746 16372010 22933657
EMBL
CT827859
GU455413
AABR07072397
AC114452
ADV57650.1
CR858584
+ More
CR858606 AK145298 BC039916 GU455414 AK022614 AK024318 AK296493 AK299883 AK302438 CH471057 BC037574 AK006739 AK035629 AK167153 BC049274 BC055398 BC068136 AF441835 BT030596 AF022789 AK289685 AL158062 AL355473 CH471075 BC026072 BC115286 BC094130 BC087530 BT044911 BT059552 Z29095 U76846 AC007168 CP002685 AY048261 AY072625 AY087731 U76845 AL035708 AL161596 CP002687 AY058889 AY079044 LLXK01000160 PKY16949.1 QKKE01000224 RGB33822.1 BDIQ01000067 AUPC02000034 GBC23597.1 POG78215.1 LLXJ01000091 LLXH01000206 PKC15367.1 PKC70685.1 KK207915 EZF48942.1 GL882890 EGF77996.1 MSZS01000001 PKX98674.1 KV424657 KZV64218.1 DS547091 EDR15274.1 KL198006 KDQ31360.1 JH971385 EKM83950.1 NHYE01005668 PPQ64280.1 KN831961 KIO06970.1 BN001304 CBF79002.1 LXFE01000603 OLL24819.1 KV417501 KZP28893.1 KQ962072 KXN90105.1 LUGH01000052 OBZ90357.1 LLXL01000022 PKK80130.1 CDGI01000810 CEJ02685.1 KZ151540 OUM56960.1 BEXD01004073 GBC06426.1 CCYT01000580 CEG83784.1 KZ819605 PWN32411.1 CEJ02683.1 CEJ02684.1 PJQL01000191 RCH98522.1 CCYT01000013 CEG69609.1 PJQL01002201 RCH85032.1 NBII01000002 PAV22641.1 CH476741 EIE87684.1 KE124014 EPB85328.1 NHTK01005754 PPQ74417.1 MCGE01000016 ORZ13725.1 CEG83783.1 LK023357 CDS12439.1 KV921438 ORE15118.1 CR382129 CAG82386.1 KV700153 OCF30351.1 EQ966262 EED83732.1 LN733222 CEP16668.1 MCGE01000003 ORZ23182.1 CBTN010000077 CDH59889.1 AEIJ01000476 GL541699 KDE04770.1 MCGN01000006 ORY95321.1 FJUW01000008 CZS94450.1 AJIL01000061 KNE98063.1 KN881675 KIY50480.1 KN840541 KIP05525.1 LLXI01000011 PKY37830.1 LN679119 CEL56060.1 KV922005 ORE03397.1 JATN01000289 EUC67048.1 LAVV01009790 KNZ49979.1 HG529615 CDI54449.1 RBNK01001468 RUS19216.1 JH658470 EXK79473.1 ANFO01000734 KGQ07043.1 KV425900 KZW00495.1 GL636488 EFW20689.1 AMGY01000009 EXJ78362.1 BEXD01001817 GBB95876.1 QKRW01000002 RAL67842.1 DS990636 EGC41364.1 GG692419 EER45423.1 QWIN01000007 RMY62737.1 PYWC01000111 PWW72274.1 CH476615 EEP75776.1 LYCR01000027 OGM46912.1 KZ107843 OSS49774.1 KV407454 KZF26009.1 KL197710 KDQ62992.1 QWIQ01000293 RMY96198.1 MCFG01000425 ORX68440.1 QKRW01000001 RAL68428.1 CH476597 JZDT01000752 KOC16469.1 FP929127 CBX95574.1 AMWN01000004 EXJ88307.1 LUGG01000034 OBZ66035.1 JZEE01000622 KJK62453.1 ML120565 RPA89611.1 AP007150 LT854253 SMR41623.1 AKHY01000215 EIT72488.1
CR858606 AK145298 BC039916 GU455414 AK022614 AK024318 AK296493 AK299883 AK302438 CH471057 BC037574 AK006739 AK035629 AK167153 BC049274 BC055398 BC068136 AF441835 BT030596 AF022789 AK289685 AL158062 AL355473 CH471075 BC026072 BC115286 BC094130 BC087530 BT044911 BT059552 Z29095 U76846 AC007168 CP002685 AY048261 AY072625 AY087731 U76845 AL035708 AL161596 CP002687 AY058889 AY079044 LLXK01000160 PKY16949.1 QKKE01000224 RGB33822.1 BDIQ01000067 AUPC02000034 GBC23597.1 POG78215.1 LLXJ01000091 LLXH01000206 PKC15367.1 PKC70685.1 KK207915 EZF48942.1 GL882890 EGF77996.1 MSZS01000001 PKX98674.1 KV424657 KZV64218.1 DS547091 EDR15274.1 KL198006 KDQ31360.1 JH971385 EKM83950.1 NHYE01005668 PPQ64280.1 KN831961 KIO06970.1 BN001304 CBF79002.1 LXFE01000603 OLL24819.1 KV417501 KZP28893.1 KQ962072 KXN90105.1 LUGH01000052 OBZ90357.1 LLXL01000022 PKK80130.1 CDGI01000810 CEJ02685.1 KZ151540 OUM56960.1 BEXD01004073 GBC06426.1 CCYT01000580 CEG83784.1 KZ819605 PWN32411.1 CEJ02683.1 CEJ02684.1 PJQL01000191 RCH98522.1 CCYT01000013 CEG69609.1 PJQL01002201 RCH85032.1 NBII01000002 PAV22641.1 CH476741 EIE87684.1 KE124014 EPB85328.1 NHTK01005754 PPQ74417.1 MCGE01000016 ORZ13725.1 CEG83783.1 LK023357 CDS12439.1 KV921438 ORE15118.1 CR382129 CAG82386.1 KV700153 OCF30351.1 EQ966262 EED83732.1 LN733222 CEP16668.1 MCGE01000003 ORZ23182.1 CBTN010000077 CDH59889.1 AEIJ01000476 GL541699 KDE04770.1 MCGN01000006 ORY95321.1 FJUW01000008 CZS94450.1 AJIL01000061 KNE98063.1 KN881675 KIY50480.1 KN840541 KIP05525.1 LLXI01000011 PKY37830.1 LN679119 CEL56060.1 KV922005 ORE03397.1 JATN01000289 EUC67048.1 LAVV01009790 KNZ49979.1 HG529615 CDI54449.1 RBNK01001468 RUS19216.1 JH658470 EXK79473.1 ANFO01000734 KGQ07043.1 KV425900 KZW00495.1 GL636488 EFW20689.1 AMGY01000009 EXJ78362.1 BEXD01001817 GBB95876.1 QKRW01000002 RAL67842.1 DS990636 EGC41364.1 GG692419 EER45423.1 QWIN01000007 RMY62737.1 PYWC01000111 PWW72274.1 CH476615 EEP75776.1 LYCR01000027 OGM46912.1 KZ107843 OSS49774.1 KV407454 KZF26009.1 KL197710 KDQ62992.1 QWIQ01000293 RMY96198.1 MCFG01000425 ORX68440.1 QKRW01000001 RAL68428.1 CH476597 JZDT01000752 KOC16469.1 FP929127 CBX95574.1 AMWN01000004 EXJ88307.1 LUGG01000034 OBZ66035.1 JZEE01000622 KJK62453.1 ML120565 RPA89611.1 AP007150 LT854253 SMR41623.1 AKHY01000215 EIT72488.1
Proteomes
UP000000437
UP000002494
UP000001595
UP000000589
UP000005640
UP000009136
+ More
UP000087266 UP000001940 UP000006548 UP000263633 UP000018888 UP000236242 UP000232688 UP000232722 UP000023758 UP000007241 UP000234474 UP000077086 UP000001194 UP000027073 UP000008493 UP000284706 UP000054217 UP000000560 UP000186594 UP000076532 UP000070249 UP000093000 UP000233469 UP000038169 UP000195826 UP000247702 UP000054919 UP000245771 UP000252139 UP000217199 UP000009138 UP000014254 UP000284842 UP000193560 UP000242381 UP000001300 UP000092666 UP000001743 UP000054107 UP000027586 UP000017200 UP000242180 UP000178129 UP000054564 UP000053257 UP000234323 UP000059188 UP000242414 UP000030108 UP000037035 UP000030663 UP000030106 UP000077266 UP000002497 UP000019478 UP000008142 UP000002624 UP000270230 UP000246991 UP000002058 UP000179179 UP000193240 UP000076632 UP000027265 UP000281468 UP000193944 UP000007963 UP000032444 UP000002668 UP000019484 UP000092993 UP000033540 UP000276215 UP000006564 UP000245764 UP000002812
UP000087266 UP000001940 UP000006548 UP000263633 UP000018888 UP000236242 UP000232688 UP000232722 UP000023758 UP000007241 UP000234474 UP000077086 UP000001194 UP000027073 UP000008493 UP000284706 UP000054217 UP000000560 UP000186594 UP000076532 UP000070249 UP000093000 UP000233469 UP000038169 UP000195826 UP000247702 UP000054919 UP000245771 UP000252139 UP000217199 UP000009138 UP000014254 UP000284842 UP000193560 UP000242381 UP000001300 UP000092666 UP000001743 UP000054107 UP000027586 UP000017200 UP000242180 UP000178129 UP000054564 UP000053257 UP000234323 UP000059188 UP000242414 UP000030108 UP000037035 UP000030663 UP000030106 UP000077266 UP000002497 UP000019478 UP000008142 UP000002624 UP000270230 UP000246991 UP000002058 UP000179179 UP000193240 UP000076632 UP000027265 UP000281468 UP000193944 UP000007963 UP000032444 UP000002668 UP000019484 UP000092993 UP000033540 UP000276215 UP000006564 UP000245764 UP000002812
PRIDE
Pfam
PF00443 UCH
+ More
PF08174 Anillin
PF01979 Amidohydro_1
PF00780 CNH
PF15405 PH_5
PF00069 Pkinase
PF03473 MOSC
PF03476 MOSC_N
PF01425 Amidase
PF13230 GATase_4
PF12022 DUF3510
PF06148 COG2
PF00503 G-alpha
PF00618 RasGEF_N
PF00617 RasGEF
PF02518 HATPase_c
PF00072 Response_reg
PF13185 GAF_2
PF13191 AAA_16
PF00512 HisKA
PF00722 Glyco_hydro_16
PF07690 MFS_1
PF00887 ACBP
PF01529 DHHC
PF08174 Anillin
PF01979 Amidohydro_1
PF00780 CNH
PF15405 PH_5
PF00069 Pkinase
PF03473 MOSC
PF03476 MOSC_N
PF01425 Amidase
PF13230 GATase_4
PF12022 DUF3510
PF06148 COG2
PF00503 G-alpha
PF00618 RasGEF_N
PF00617 RasGEF
PF02518 HATPase_c
PF00072 Response_reg
PF13185 GAF_2
PF13191 AAA_16
PF00512 HisKA
PF00722 Glyco_hydro_16
PF07690 MFS_1
PF00887 ACBP
PF01529 DHHC
Interpro
IPR001394
Peptidase_C19_UCH
+ More
IPR018200 USP_CS
IPR038765 Papain-like_cys_pep_sf
IPR028889 USP_dom
IPR034261 CNOT4_RRM
IPR013083 Znf_RING/FYVE/PHD
IPR039515 NOT4_mRING-HC-C4C4
IPR003954 RRM_dom_euk
IPR035979 RBD_domain_sf
IPR001841 Znf_RING
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR039780 Mot2
IPR012966 AHD
IPR032466 Metal_Hydrolase
IPR011059 Metal-dep_hydrolase_composite
IPR006680 Amidohydro-rel
IPR001180 CNH_dom
IPR001849 PH_domain
IPR011993 PH-like_dom_sf
IPR041675 PH_5
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR005303 MOSC_N
IPR005302 MoCF_Sase_C
IPR023631 Amidase_dom
IPR000120 Amidase
IPR036928 AS_sf
IPR026869 Put_GATase_2
IPR029055 Ntn_hydrolases_N
IPR017932 GATase_2_dom
IPR024602 COG_su2_N
IPR024603 COG_complex_COG2_C
IPR009316 COG2
IPR027417 P-loop_NTPase
IPR011025 GproteinA_insert
IPR001019 Gprotein_alpha_su
IPR001895 RASGEF_cat_dom
IPR036964 RASGEF_cat_dom_sf
IPR008937 Ras-like_GEF
IPR023578 Ras_GEF_dom_sf
IPR000651 Ras-like_Gua-exchang_fac_N
IPR004358 Sig_transdc_His_kin-like_C
IPR003594 HATPase_C
IPR036097 HisK_dim/P_sf
IPR001789 Sig_transdc_resp-reg_receiver
IPR036890 HATPase_C_sf
IPR041664 AAA_16
IPR003661 HisK_dim/P
IPR029016 GAF-like_dom_sf
IPR011006 CheY-like_superfamily
IPR003018 GAF
IPR005467 His_kinase_dom
IPR013320 ConA-like_dom_sf
IPR000757 GH16
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR000582 Acyl-CoA-binding_protein
IPR035984 Acyl-CoA-binding_sf
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR001594 Palmitoyltrfase_DHHC
IPR018200 USP_CS
IPR038765 Papain-like_cys_pep_sf
IPR028889 USP_dom
IPR034261 CNOT4_RRM
IPR013083 Znf_RING/FYVE/PHD
IPR039515 NOT4_mRING-HC-C4C4
IPR003954 RRM_dom_euk
IPR035979 RBD_domain_sf
IPR001841 Znf_RING
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR039780 Mot2
IPR012966 AHD
IPR032466 Metal_Hydrolase
IPR011059 Metal-dep_hydrolase_composite
IPR006680 Amidohydro-rel
IPR001180 CNH_dom
IPR001849 PH_domain
IPR011993 PH-like_dom_sf
IPR041675 PH_5
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR005303 MOSC_N
IPR005302 MoCF_Sase_C
IPR023631 Amidase_dom
IPR000120 Amidase
IPR036928 AS_sf
IPR026869 Put_GATase_2
IPR029055 Ntn_hydrolases_N
IPR017932 GATase_2_dom
IPR024602 COG_su2_N
IPR024603 COG_complex_COG2_C
IPR009316 COG2
IPR027417 P-loop_NTPase
IPR011025 GproteinA_insert
IPR001019 Gprotein_alpha_su
IPR001895 RASGEF_cat_dom
IPR036964 RASGEF_cat_dom_sf
IPR008937 Ras-like_GEF
IPR023578 Ras_GEF_dom_sf
IPR000651 Ras-like_Gua-exchang_fac_N
IPR004358 Sig_transdc_His_kin-like_C
IPR003594 HATPase_C
IPR036097 HisK_dim/P_sf
IPR001789 Sig_transdc_resp-reg_receiver
IPR036890 HATPase_C_sf
IPR041664 AAA_16
IPR003661 HisK_dim/P
IPR029016 GAF-like_dom_sf
IPR011006 CheY-like_superfamily
IPR003018 GAF
IPR005467 His_kinase_dom
IPR013320 ConA-like_dom_sf
IPR000757 GH16
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR000582 Acyl-CoA-binding_protein
IPR035984 Acyl-CoA-binding_sf
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR001594 Palmitoyltrfase_DHHC
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A5WWB0
F1M625
Q5RBQ4
P62069
P62068
Q9D9M2
+ More
A5D9H7 O75317 A4FUN7 Q52KZ6 Q5M981 C0HB46 P34547 Q8LAM0 O24454 A0A2I1E4B4 A0A372R1S4 A0A2H5RZT2 A0A2N0S567 A0A022VRL4 F4P9S8 A0A2I1CM32 A0A166ATA3 B0CNB1 A0A067NWK7 K5Y6W7 A0A409VEF5 A0A0C3P1N6 C8VD90 A0A1U7LQB1 A0A166S0Z9 A0A137QS67 A0A1C7NMP4 A0A2N1P1T7 A0A0A1PD09 A0A1Y3MMV9 A0A2Z6RTU0 A0A0C7CJW2 A0A316V7D4 A0A0A1NTD1 A0A0A1PF16 A0A367K8G3 A0A0C7BF14 A0A367J515 A0A286UST7 I1CGV4 S2JRR9 A0A409W7B0 A0A1X2ICC7 A0A0C7CLL8 A0A077WYX3 A0A1X0RTB5 Q6CB96 A0A1B9GHD3 B8P5U4 A0A0B7NMT2 A0A1X2IVZ3 A0A068SC62 U5HCE2 A0A1X2H9H2 A0A1E1K971 A0A0L0VFN5 A0A0D7AJH2 A0A0C3S8P8 A0A2I1FTX9 A0A0B7FEQ6 A0A1X0QUH1 X8JVI7 A0A0L6UN82 A0A077RB21 A0A433PNY3 X0BKP4 A0A0A2VHI9 A0A165NBC2 E9CXL2 W9XD26 A0A2Z6RV19 A0A395J5Q6 F0U5R9 C6H2S0 A0A3M7DFA1 A0A317SD19 C4JKS7 A0A1F8A596 A0A1Y2M0T9 A0A165JBF0 A0A067QHM1 A0A3M7G508 A0A1Y1W4E6 A0A395J8G9 Q0CT11 A0A2P2HNM3 E4ZVE8 W9Y652 A0A1C7LMM6 A0A0F0I917 A0A3N4IZ82 Q2UUG8 A0A2H1FJX3 I8TFM0
A5D9H7 O75317 A4FUN7 Q52KZ6 Q5M981 C0HB46 P34547 Q8LAM0 O24454 A0A2I1E4B4 A0A372R1S4 A0A2H5RZT2 A0A2N0S567 A0A022VRL4 F4P9S8 A0A2I1CM32 A0A166ATA3 B0CNB1 A0A067NWK7 K5Y6W7 A0A409VEF5 A0A0C3P1N6 C8VD90 A0A1U7LQB1 A0A166S0Z9 A0A137QS67 A0A1C7NMP4 A0A2N1P1T7 A0A0A1PD09 A0A1Y3MMV9 A0A2Z6RTU0 A0A0C7CJW2 A0A316V7D4 A0A0A1NTD1 A0A0A1PF16 A0A367K8G3 A0A0C7BF14 A0A367J515 A0A286UST7 I1CGV4 S2JRR9 A0A409W7B0 A0A1X2ICC7 A0A0C7CLL8 A0A077WYX3 A0A1X0RTB5 Q6CB96 A0A1B9GHD3 B8P5U4 A0A0B7NMT2 A0A1X2IVZ3 A0A068SC62 U5HCE2 A0A1X2H9H2 A0A1E1K971 A0A0L0VFN5 A0A0D7AJH2 A0A0C3S8P8 A0A2I1FTX9 A0A0B7FEQ6 A0A1X0QUH1 X8JVI7 A0A0L6UN82 A0A077RB21 A0A433PNY3 X0BKP4 A0A0A2VHI9 A0A165NBC2 E9CXL2 W9XD26 A0A2Z6RV19 A0A395J5Q6 F0U5R9 C6H2S0 A0A3M7DFA1 A0A317SD19 C4JKS7 A0A1F8A596 A0A1Y2M0T9 A0A165JBF0 A0A067QHM1 A0A3M7G508 A0A1Y1W4E6 A0A395J8G9 Q0CT11 A0A2P2HNM3 E4ZVE8 W9Y652 A0A1C7LMM6 A0A0F0I917 A0A3N4IZ82 Q2UUG8 A0A2H1FJX3 I8TFM0
PDB
5L8W
E-value=1.48795e-153,
Score=1392
Ontologies
GO
GO:0006511
GO:0007610
GO:0004197
GO:0046872
GO:0032228
GO:0016579
GO:0004843
GO:0036459
GO:0001662
GO:0008343
GO:0098978
GO:0045202
GO:0099149
GO:0060013
GO:0048149
GO:0050862
GO:0005829
GO:0101005
GO:0005634
GO:0005654
GO:0090326
GO:2000311
GO:0005737
GO:0043025
GO:0010628
GO:0005768
GO:0008595
GO:0043204
GO:0035255
GO:0071947
GO:0003723
GO:0004842
GO:0030014
GO:0006147
GO:0046098
GO:0008270
GO:0008892
GO:0005524
GO:0004672
GO:0030151
GO:0030170
GO:0004040
GO:0016853
GO:0016740
GO:0007030
GO:0016020
GO:0015031
GO:0031683
GO:0003924
GO:0007186
GO:0005525
GO:0016021
GO:0007264
GO:0005085
GO:0000155
GO:0005975
GO:0004553
GO:0019706
GO:0045013
GO:0055085
GO:0008061
GO:0006412
GO:0015934
GO:0015914
GO:0016876
GO:0043039
Topology
Subcellular location
Perikaryon
Cytoplasm
Nucleus
Cytoplasm
Nucleus
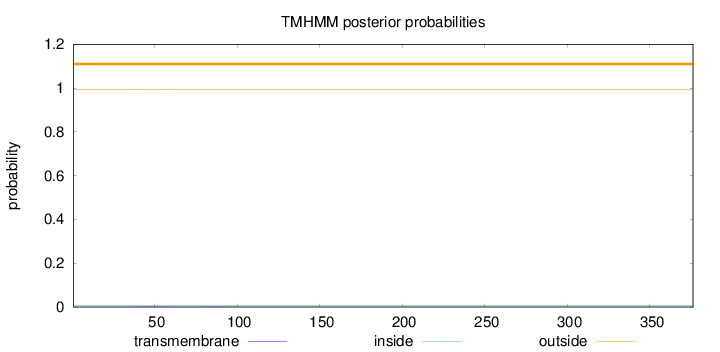
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0320500000000001
Exp number, first 60 AAs:
0.02854
Total prob of N-in:
0.00623
outside
1 - 376
Population Genetic Test Statistics
Pi
2.438376
Theta
21.269544
Tajima's D
-0.849459
CLR
2.031511
CSRT
0.165641717914104
Interpretation
Uncertain
