Gene
KWMTBOMO08097 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001107
Annotation
DNA_supercoiling_factor_[Bombyx_mori]
Full name
Calumenin-B
+ More
Calumenin
Calumenin
Alternative Name
Crocalbin
IEF SSP 9302
IEF SSP 9302
Location in the cell
Cytoplasmic Reliability : 2.085
Sequence
CDS
ATGTACAAACCCGAGGACGGCGAGGACGAGGAGGAACCCGACTGGGTCAAGCAGGAGAGAGAACAGTTTACTGGATACAGAGATACGAATAAGGACGGGTTCATGGACGAGCACGAGGTGAAGGACTGGATCGCGCCGCCGGAGTTCGACCACGCGGAGGCCGAGGCGCGCCACCTCGTGTTCGAGGCCGACGCCGACGCAGACGAGAAGCTCACCAAGGCCGAGATCATCGACAAGTACGATCTCTTCGTCGGCTCGCAGGCCACCGACTTCGGCGAAGCACTCGCCAGACACGACGAATTTTAA
Protein
MYKPEDGEDEEEPDWVKQEREQFTGYRDTNKDGFMDEHEVKDWIAPPEFDHAEAEARHLVFEADADADEKLTKAEIIDKYDLFVGSQATDFGEALARHDEF
Summary
Description
Involved in regulation of vitamin K-dependent carboxylation of multiple N-terminal glutamate residues. Seems to inhibit gamma-carboxylase ggcx. Binds 7 calcium ions with a low affinity (By similarity).
Involved in regulation of vitamin K-dependent carboxylation of multiple N-terminal glutamate residues. Seems to inhibit gamma-carboxylase GGCX. Binds 7 calcium ions with a low affinity (By similarity).
Involved in regulation of vitamin K-dependent carboxylation of multiple N-terminal glutamate residues. Seems to inhibit gamma-carboxylase GGCX. Binds 7 calcium ions with a low affinity (By similarity).
Subunit
Interacts with ggcx.
Interacts with GGCX.
Interacts with GGCX.
Similarity
Belongs to the CREC family.
Keywords
Calcium
Complete proteome
Endoplasmic reticulum
Glycoprotein
Golgi apparatus
Membrane
Metal-binding
Reference proteome
Repeat
Sarcoplasmic reticulum
Secreted
Signal
Acetylation
Alternative splicing
Direct protein sequencing
Phosphoprotein
Polymorphism
Feature
chain Calumenin-B
splice variant In isoform 15.
sequence variant In dbSNP:rs2290228.
splice variant In isoform 15.
sequence variant In dbSNP:rs2290228.
Uniprot
G9FL14
Q93136
A0A194RF50
A0A194Q7Q8
A0A2A4JW54
A0A2H1WQL7
+ More
S4PSB2 A0A212EX33 A0A3S2PK98 A0A182U3Q0 A0A182XFN2 A0A182LN58 Q7QDS4 A0A182HX52 A0A182MFS9 A0A182VP64 A0A182K9R5 A0A182SIW5 A0A182YE68 A0A182P1A0 A0A182R8X5 A0A0T6B3A3 E0VBH5 A0A182NVC3 A0A182Q3Y8 A0A182W8B6 A0A182FLY7 A0A2M4BWV7 D2A169 A0A2M4BUB1 W5JBJ4 A0A084VJI4 A0A182J1Q4 A0A2J7Q139 A0A023EQT3 A0A2J7Q112 A0A2M4AWK1 Q0IEY6 V9PBR3 A0A1S4EKS1 B0W572 Q207I9 A0A1Q3FMF4 A0A1Y1K6Q0 A0A1Q3FML7 A0A1B6JUC7 A0A154PNC9 A0A1B6HBB1 A0A1S3ML90 B5X4E0 T1JLL4 A0A1B0DFV8 A0A1W7R9T9 A0A060XGU7 A0A0C9RE82 A0A0L7QY92 A0A3B4FM91 A0A3Q2WW28 A0A195B6F8 F4WSS6 A0A2J8QT00 H0Y875 A0A3Q3LJU3 I3KCN5 A0A195FPW8 A0A151XD84 A0A1B6E6K0 A0A158NH09 O43852-15 A0A2R2MS28 A0A250YAM7 A0A1B6MN73 G3V004 A0A067RB55 A0A3B3IHZ0 A0A3P9LP03 A0A3B3D771 A0A3P9I4F0 A0A3P9LNU5 A0A151J1Y5 O96051 A0A0C9R530 B3KQK3 A0A2G9S1D1 A0A3P9AHZ4 G3PH64 C1BTZ9 A0A3P8R1F8 A0A3P9CWS2 G3PH67 A0A3P8UT21 A0A3Q1H6T2 C3ZK54 Q8IRH7 M3ZYR1 A0A2U9BW86 A0A315US90 A0A3B4YA47 A0A3B4UIB0 A0A3B5MUN6
S4PSB2 A0A212EX33 A0A3S2PK98 A0A182U3Q0 A0A182XFN2 A0A182LN58 Q7QDS4 A0A182HX52 A0A182MFS9 A0A182VP64 A0A182K9R5 A0A182SIW5 A0A182YE68 A0A182P1A0 A0A182R8X5 A0A0T6B3A3 E0VBH5 A0A182NVC3 A0A182Q3Y8 A0A182W8B6 A0A182FLY7 A0A2M4BWV7 D2A169 A0A2M4BUB1 W5JBJ4 A0A084VJI4 A0A182J1Q4 A0A2J7Q139 A0A023EQT3 A0A2J7Q112 A0A2M4AWK1 Q0IEY6 V9PBR3 A0A1S4EKS1 B0W572 Q207I9 A0A1Q3FMF4 A0A1Y1K6Q0 A0A1Q3FML7 A0A1B6JUC7 A0A154PNC9 A0A1B6HBB1 A0A1S3ML90 B5X4E0 T1JLL4 A0A1B0DFV8 A0A1W7R9T9 A0A060XGU7 A0A0C9RE82 A0A0L7QY92 A0A3B4FM91 A0A3Q2WW28 A0A195B6F8 F4WSS6 A0A2J8QT00 H0Y875 A0A3Q3LJU3 I3KCN5 A0A195FPW8 A0A151XD84 A0A1B6E6K0 A0A158NH09 O43852-15 A0A2R2MS28 A0A250YAM7 A0A1B6MN73 G3V004 A0A067RB55 A0A3B3IHZ0 A0A3P9LP03 A0A3B3D771 A0A3P9I4F0 A0A3P9LNU5 A0A151J1Y5 O96051 A0A0C9R530 B3KQK3 A0A2G9S1D1 A0A3P9AHZ4 G3PH64 C1BTZ9 A0A3P8R1F8 A0A3P9CWS2 G3PH67 A0A3P8UT21 A0A3Q1H6T2 C3ZK54 Q8IRH7 M3ZYR1 A0A2U9BW86 A0A315US90 A0A3B4YA47 A0A3B4UIB0 A0A3B5MUN6
Pubmed
19121390
7797553
26354079
23622113
22118469
20966253
+ More
12364791 14747013 17210077 25244985 20566863 18362917 19820115 20920257 23761445 24438588 24945155 26483478 17510324 28004739 20433749 24755649 21719571 12853948 21269460 23186163 25944712 25186727 21347285 9675259 9598325 14702039 15489334 12665801 10222138 12643545 17081065 16964243 18691976 19159218 19139490 20068231 24275569 26091039 28087693 19468303 21183079 23806337 24845553 17554307 29451363 9774687 16303743 25069045 24487278 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23542700 29703783
12364791 14747013 17210077 25244985 20566863 18362917 19820115 20920257 23761445 24438588 24945155 26483478 17510324 28004739 20433749 24755649 21719571 12853948 21269460 23186163 25944712 25186727 21347285 9675259 9598325 14702039 15489334 12665801 10222138 12643545 17081065 16964243 18691976 19159218 19139490 20068231 24275569 26091039 28087693 19468303 21183079 23806337 24845553 17554307 29451363 9774687 16303743 25069045 24487278 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23542700 29703783
EMBL
BABH01000173
JN171664
AEO79985.1
D49948
BAA08704.1
KQ460297
+ More
KPJ16212.1 KQ459439 KPJ01030.1 NWSH01000457 PCG76267.1 ODYU01009967 SOQ54734.1 GAIX01014008 JAA78552.1 AGBW02011840 OWR46049.1 RSAL01000009 RVE53846.1 AAAB01008849 EAA07240.3 APCN01005137 AXCM01001696 LJIG01016193 KRT81335.1 DS235030 EEB10731.1 AXCN02002295 GGFJ01008067 MBW57208.1 KQ971338 EFA02623.1 GGFJ01007390 MBW56531.1 ADMH02001606 ETN61842.1 ATLV01013581 KE524868 KFB38128.1 AXCP01007332 NEVH01019960 PNF22279.1 JXUM01101857 JXUM01101858 GAPW01002619 JAC10979.1 PNF22278.1 GGFK01011856 MBW45177.1 CH477461 EAT40554.1 KC282397 AGX25154.1 DS231841 EDS34819.1 DQ407909 ABD65581.1 GFDL01006320 JAV28725.1 GEZM01091041 GEZM01091040 JAV57132.1 GFDL01006319 JAV28726.1 GECU01033021 GECU01027847 GECU01004891 GECU01004505 JAS74685.1 JAS79859.1 JAT02816.1 JAT03202.1 KQ434996 KZC13237.1 GECU01035715 GECU01035055 GECU01034243 GECU01024945 GECU01021570 GECU01021011 GECU01005931 JAS71991.1 JAS72651.1 JAS73463.1 JAS82761.1 JAS86136.1 JAS86695.1 JAT01776.1 BT045909 JH432114 AJVK01059049 GFAH01000485 JAV47904.1 FR905356 CDQ78671.1 GBYB01011462 JAG81229.1 KQ414693 KOC63572.1 KQ976574 KYM80103.1 GL888327 EGI62733.1 NBAG03000017 PNI99406.1 AC024952 AERX01005254 KQ981387 KYN41979.1 KQ982294 KYQ58334.1 GEDC01031670 GEDC01029371 GEDC01017660 GEDC01004022 GEDC01003728 GEDC01001923 JAS05628.1 JAS07927.1 JAS19638.1 JAS33276.1 JAS33570.1 JAS35375.1 ADTU01015424 U67280 AF013759 AF345637 HM002604 HM002605 HM002606 HM002607 HM002608 HM002609 HM002610 HM002611 HM002612 HM002613 HM002614 HM002615 HM002616 AK056338 CR541835 CH471070 BC013383 AF257659 GFFW01004096 JAV40692.1 GEBQ01002658 JAT37319.1 AC044807 KK852761 KDR16997.1 KQ980447 KYN16001.1 AB011261 BAA34049.1 GBYB01011424 JAG81191.1 AK075089 BAG52065.1 KV927880 PIO33959.1 BT078078 ACO12502.1 GG666636 EEN46953.1 AE014296 JQ663526 AAN11468.1 AFI26260.1 CP026252 AWP08547.1 NHOQ01002850 PWA14215.1
KPJ16212.1 KQ459439 KPJ01030.1 NWSH01000457 PCG76267.1 ODYU01009967 SOQ54734.1 GAIX01014008 JAA78552.1 AGBW02011840 OWR46049.1 RSAL01000009 RVE53846.1 AAAB01008849 EAA07240.3 APCN01005137 AXCM01001696 LJIG01016193 KRT81335.1 DS235030 EEB10731.1 AXCN02002295 GGFJ01008067 MBW57208.1 KQ971338 EFA02623.1 GGFJ01007390 MBW56531.1 ADMH02001606 ETN61842.1 ATLV01013581 KE524868 KFB38128.1 AXCP01007332 NEVH01019960 PNF22279.1 JXUM01101857 JXUM01101858 GAPW01002619 JAC10979.1 PNF22278.1 GGFK01011856 MBW45177.1 CH477461 EAT40554.1 KC282397 AGX25154.1 DS231841 EDS34819.1 DQ407909 ABD65581.1 GFDL01006320 JAV28725.1 GEZM01091041 GEZM01091040 JAV57132.1 GFDL01006319 JAV28726.1 GECU01033021 GECU01027847 GECU01004891 GECU01004505 JAS74685.1 JAS79859.1 JAT02816.1 JAT03202.1 KQ434996 KZC13237.1 GECU01035715 GECU01035055 GECU01034243 GECU01024945 GECU01021570 GECU01021011 GECU01005931 JAS71991.1 JAS72651.1 JAS73463.1 JAS82761.1 JAS86136.1 JAS86695.1 JAT01776.1 BT045909 JH432114 AJVK01059049 GFAH01000485 JAV47904.1 FR905356 CDQ78671.1 GBYB01011462 JAG81229.1 KQ414693 KOC63572.1 KQ976574 KYM80103.1 GL888327 EGI62733.1 NBAG03000017 PNI99406.1 AC024952 AERX01005254 KQ981387 KYN41979.1 KQ982294 KYQ58334.1 GEDC01031670 GEDC01029371 GEDC01017660 GEDC01004022 GEDC01003728 GEDC01001923 JAS05628.1 JAS07927.1 JAS19638.1 JAS33276.1 JAS33570.1 JAS35375.1 ADTU01015424 U67280 AF013759 AF345637 HM002604 HM002605 HM002606 HM002607 HM002608 HM002609 HM002610 HM002611 HM002612 HM002613 HM002614 HM002615 HM002616 AK056338 CR541835 CH471070 BC013383 AF257659 GFFW01004096 JAV40692.1 GEBQ01002658 JAT37319.1 AC044807 KK852761 KDR16997.1 KQ980447 KYN16001.1 AB011261 BAA34049.1 GBYB01011424 JAG81191.1 AK075089 BAG52065.1 KV927880 PIO33959.1 BT078078 ACO12502.1 GG666636 EEN46953.1 AE014296 JQ663526 AAN11468.1 AFI26260.1 CP026252 AWP08547.1 NHOQ01002850 PWA14215.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
+ More
UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075883 UP000075903 UP000075881 UP000075901 UP000076408 UP000075885 UP000075900 UP000009046 UP000075884 UP000075886 UP000075920 UP000069272 UP000007266 UP000000673 UP000030765 UP000075880 UP000235965 UP000069940 UP000008820 UP000079169 UP000002320 UP000076502 UP000087266 UP000092462 UP000193380 UP000053825 UP000261460 UP000264840 UP000078540 UP000007755 UP000005640 UP000261640 UP000005207 UP000078541 UP000075809 UP000005205 UP000085678 UP000000589 UP000027135 UP000001038 UP000265180 UP000261560 UP000265200 UP000078492 UP000265140 UP000007635 UP000265100 UP000265160 UP000265120 UP000265040 UP000001554 UP000000803 UP000002852 UP000246464 UP000261360 UP000261420 UP000261380
UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075883 UP000075903 UP000075881 UP000075901 UP000076408 UP000075885 UP000075900 UP000009046 UP000075884 UP000075886 UP000075920 UP000069272 UP000007266 UP000000673 UP000030765 UP000075880 UP000235965 UP000069940 UP000008820 UP000079169 UP000002320 UP000076502 UP000087266 UP000092462 UP000193380 UP000053825 UP000261460 UP000264840 UP000078540 UP000007755 UP000005640 UP000261640 UP000005207 UP000078541 UP000075809 UP000005205 UP000085678 UP000000589 UP000027135 UP000001038 UP000265180 UP000261560 UP000265200 UP000078492 UP000265140 UP000007635 UP000265100 UP000265160 UP000265120 UP000265040 UP000001554 UP000000803 UP000002852 UP000246464 UP000261360 UP000261420 UP000261380
Interpro
Gene 3D
ProteinModelPortal
G9FL14
Q93136
A0A194RF50
A0A194Q7Q8
A0A2A4JW54
A0A2H1WQL7
+ More
S4PSB2 A0A212EX33 A0A3S2PK98 A0A182U3Q0 A0A182XFN2 A0A182LN58 Q7QDS4 A0A182HX52 A0A182MFS9 A0A182VP64 A0A182K9R5 A0A182SIW5 A0A182YE68 A0A182P1A0 A0A182R8X5 A0A0T6B3A3 E0VBH5 A0A182NVC3 A0A182Q3Y8 A0A182W8B6 A0A182FLY7 A0A2M4BWV7 D2A169 A0A2M4BUB1 W5JBJ4 A0A084VJI4 A0A182J1Q4 A0A2J7Q139 A0A023EQT3 A0A2J7Q112 A0A2M4AWK1 Q0IEY6 V9PBR3 A0A1S4EKS1 B0W572 Q207I9 A0A1Q3FMF4 A0A1Y1K6Q0 A0A1Q3FML7 A0A1B6JUC7 A0A154PNC9 A0A1B6HBB1 A0A1S3ML90 B5X4E0 T1JLL4 A0A1B0DFV8 A0A1W7R9T9 A0A060XGU7 A0A0C9RE82 A0A0L7QY92 A0A3B4FM91 A0A3Q2WW28 A0A195B6F8 F4WSS6 A0A2J8QT00 H0Y875 A0A3Q3LJU3 I3KCN5 A0A195FPW8 A0A151XD84 A0A1B6E6K0 A0A158NH09 O43852-15 A0A2R2MS28 A0A250YAM7 A0A1B6MN73 G3V004 A0A067RB55 A0A3B3IHZ0 A0A3P9LP03 A0A3B3D771 A0A3P9I4F0 A0A3P9LNU5 A0A151J1Y5 O96051 A0A0C9R530 B3KQK3 A0A2G9S1D1 A0A3P9AHZ4 G3PH64 C1BTZ9 A0A3P8R1F8 A0A3P9CWS2 G3PH67 A0A3P8UT21 A0A3Q1H6T2 C3ZK54 Q8IRH7 M3ZYR1 A0A2U9BW86 A0A315US90 A0A3B4YA47 A0A3B4UIB0 A0A3B5MUN6
S4PSB2 A0A212EX33 A0A3S2PK98 A0A182U3Q0 A0A182XFN2 A0A182LN58 Q7QDS4 A0A182HX52 A0A182MFS9 A0A182VP64 A0A182K9R5 A0A182SIW5 A0A182YE68 A0A182P1A0 A0A182R8X5 A0A0T6B3A3 E0VBH5 A0A182NVC3 A0A182Q3Y8 A0A182W8B6 A0A182FLY7 A0A2M4BWV7 D2A169 A0A2M4BUB1 W5JBJ4 A0A084VJI4 A0A182J1Q4 A0A2J7Q139 A0A023EQT3 A0A2J7Q112 A0A2M4AWK1 Q0IEY6 V9PBR3 A0A1S4EKS1 B0W572 Q207I9 A0A1Q3FMF4 A0A1Y1K6Q0 A0A1Q3FML7 A0A1B6JUC7 A0A154PNC9 A0A1B6HBB1 A0A1S3ML90 B5X4E0 T1JLL4 A0A1B0DFV8 A0A1W7R9T9 A0A060XGU7 A0A0C9RE82 A0A0L7QY92 A0A3B4FM91 A0A3Q2WW28 A0A195B6F8 F4WSS6 A0A2J8QT00 H0Y875 A0A3Q3LJU3 I3KCN5 A0A195FPW8 A0A151XD84 A0A1B6E6K0 A0A158NH09 O43852-15 A0A2R2MS28 A0A250YAM7 A0A1B6MN73 G3V004 A0A067RB55 A0A3B3IHZ0 A0A3P9LP03 A0A3B3D771 A0A3P9I4F0 A0A3P9LNU5 A0A151J1Y5 O96051 A0A0C9R530 B3KQK3 A0A2G9S1D1 A0A3P9AHZ4 G3PH64 C1BTZ9 A0A3P8R1F8 A0A3P9CWS2 G3PH67 A0A3P8UT21 A0A3Q1H6T2 C3ZK54 Q8IRH7 M3ZYR1 A0A2U9BW86 A0A315US90 A0A3B4YA47 A0A3B4UIB0 A0A3B5MUN6
Ontologies
GO
PANTHER
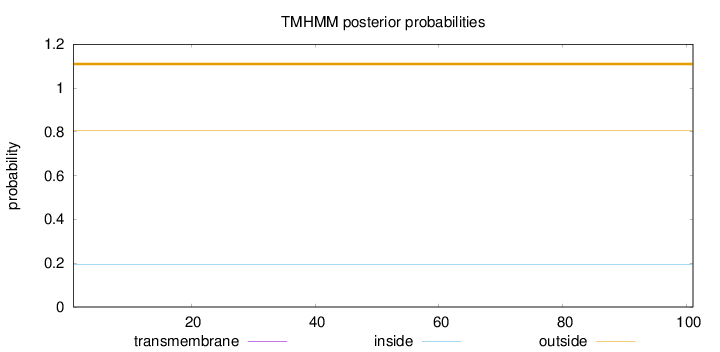
Topology
Subcellular location
Endoplasmic reticulum membrane
Golgi apparatus
Secreted
Melanosome
Sarcoplasmic reticulum lumen
Golgi apparatus
Secreted
Melanosome
Sarcoplasmic reticulum lumen
Length:
101
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.19375
outside
1 - 101
Population Genetic Test Statistics
Pi
29.780104
Theta
25.208707
Tajima's D
0.528031
CLR
0.036736
CSRT
0.524673766311684
Interpretation
Uncertain
