Gene
KWMTBOMO08072
Annotation
PREDICTED:_hemicentin-1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.8 Extracellular Reliability : 1.384
Sequence
CDS
ATGGACACACCGTCCTATGTAGCAACCGACTCTGGTGCTGCTGAGCTCACGTTTTACTATGACACTCGAGTGCTCGGGCCGTCTGCTCACTGGACTGACGAGCCCTTCGCAAACCGAGCACGCTGGTACAGCACGCCGTCTTCTGGTCTCCACGTGGATGCTGTACAGGCCCGAGACAGAGCCGTCTACAGATGCAGGGTCGATTTCCAAATCTCACCCACCAGGAACTACAGGATTGCACTTGATGTAATAGAGCTCCCGAATAAGCCGGTTATCTTTGACGAATTCGGCAAGGAAATATCCGGTGCGGCGGGCCCTTACAACGAGGGTGGGGACTTCAAACTTATTTGTTCAGTAAACGGGGGTACACCCACTCCAAGAGTACAATGGCTGCACGGAGACACAGTTTTAACGACTTTGGGAGCCAGTGACCTTCAACCTTCGGGGATGACCAGGACTCTAGTACTAGTTGTGAAGAACGCTACTAGGCTGCATTTGTCTGCTGTTTACACTTGCACGGCTGATAACACACTGTTGTCTACACCGCAGAAGACTAGCGTTCGAGTGGATCTGTATTTGAGGCCGCTATCAGTCGAAATTTTATCAAAGGAACAGCCGCTCTCCGTGGGCAGAAAGACTGAGCTTTGGTGCCGGTCTACAGGGGCACGACCCCCTGCTAATATCACTTGGTGGCTTGATGGAAAACAGCTGCACTCTATCGGCAAACAGAAAAAGGATACAAACGAAAGCCAGTCTTTGCTCGAATGGGTCCCCATGAAAGAGCACAACGGGAAGGTACTCACATGCAGAGCTGAACATGTTCGCTTCAATCACTCCACCATTGAAAGCAAGCTACCTTTGAATATTTACTACGTTCCCATTACAAGAATGGAGCTTGGCTCGAAGATGAATCCAAACGATATTGAAGAAGGCGACGATGTTTACTTCGGGTGCGACGTTGACGCAAATCCACCAGCGTACAAAGTCATATGGGAGCATAACGAGGTTCTTCTACAACACAATCCATCGAATGGTGTAATATTGACCGGGAACACGAACCTGGCGGTCCGAAACGTATCAAGACACCAAGCGGGAAACTACACTTGCACAGCATCTAACGTGGAGGGTGACGGAAAATCTCCTCCACTGCACTTACAAATAGTTTGTGAGTATACTTTAATAATGATTATTGCAGTACGTCAAGTAATAGAAATTCAATCAGTGGCACATCAAGATTTACATCCATATTAA
Protein
MDTPSYVATDSGAAELTFYYDTRVLGPSAHWTDEPFANRARWYSTPSSGLHVDAVQARDRAVYRCRVDFQISPTRNYRIALDVIELPNKPVIFDEFGKEISGAAGPYNEGGDFKLICSVNGGTPTPRVQWLHGDTVLTTLGASDLQPSGMTRTLVLVVKNATRLHLSAVYTCTADNTLLSTPQKTSVRVDLYLRPLSVEILSKEQPLSVGRKTELWCRSTGARPPANITWWLDGKQLHSIGKQKKDTNESQSLLEWVPMKEHNGKVLTCRAEHVRFNHSTIESKLPLNIYYVPITRMELGSKMNPNDIEEGDDVYFGCDVDANPPAYKVIWEHNEVLLQHNPSNGVILTGNTNLAVRNVSRHQAGNYTCTASNVEGDGKSPPLHLQIVCEYTLIMIIAVRQVIEIQSVAHQDLHPY
Summary
Uniprot
A0A212FKE7
A0A194Q659
A0A194RDG7
A0A2W1BFG7
A0A3S2P4N9
A0A437BPB1
+ More
A0A194QC59 A0A194RCY1 A0A0L7L3B9 A0A2H1WP36 K7J3W1 A0A1Y1NAB6 A0A0L7KWU5 A0A026WZS6 A0A154PQL6 A0A232FD16 A0A3L8D540 A0A195F9V8 A0A0M9ABL7 A0A195EAN5 A0A158NCS0 A0A151HZS0 A0A0C9PXU9 A0A0C9QIG5 A0A1B6DTE2 A0A0C9R081 A0A146LYE1 A0A0A9XSS5 A0A2S2NL76 A0A2H8TFI4 Q17LV3 C8VV84 A0A088AEQ8 A0A1S3D0I8 D6WPK0 A0A0L7RI46 J9K2X1 A0A1I8Q7E3 E1JII4 A0A2A3EAV1 A4IJ60 A0A0A9ZA85 A0A336LM08 A0A1S4GDS9 E0VMK9 A0A084VRP7 Q0KI85 B4HIF4 B4PLG0 B3NZK0 B4GLE2 A0A0M5IZH3 A0A182XU94 A0A1W4UT22 A0A182II24 A0A3B0KEL4 B4KAI7 W8AZH0 A0A0Q9X4G1 A0A0Q9WPR8 A0A2S2QDI2 H9J5G2 B3LZ81 A0A2A4JDY9 Q7PUJ2 A0A026WSM2 A0A3L8D7Y1 A0A158NT60 A0A154P3M9 A0A151IA00 A0A2J7PYK2 A0A212FI98 A0A182UG36 A0A194RK28 A0A437BXX4 A0A194PPB3 A0A2H1WJ89 A0A151WJA9 A0A195EGJ5 A0A2J7PYN3 A0A0P9ANK1 A0A0K8T359 A0A0A9ZBH5 C0PTV4 B0WA44 A0A067RCL4 A0A1B6G259 A0A2A3E5P9 A0A182N5E5 E0VML7 A0A182R5H8 A0A088A033 A0A212EM07 A0A1J1HQ06 A0A2R7WZ49 B3LX91 A0A1Y1NKS5 A0A1W4UR97
A0A194QC59 A0A194RCY1 A0A0L7L3B9 A0A2H1WP36 K7J3W1 A0A1Y1NAB6 A0A0L7KWU5 A0A026WZS6 A0A154PQL6 A0A232FD16 A0A3L8D540 A0A195F9V8 A0A0M9ABL7 A0A195EAN5 A0A158NCS0 A0A151HZS0 A0A0C9PXU9 A0A0C9QIG5 A0A1B6DTE2 A0A0C9R081 A0A146LYE1 A0A0A9XSS5 A0A2S2NL76 A0A2H8TFI4 Q17LV3 C8VV84 A0A088AEQ8 A0A1S3D0I8 D6WPK0 A0A0L7RI46 J9K2X1 A0A1I8Q7E3 E1JII4 A0A2A3EAV1 A4IJ60 A0A0A9ZA85 A0A336LM08 A0A1S4GDS9 E0VMK9 A0A084VRP7 Q0KI85 B4HIF4 B4PLG0 B3NZK0 B4GLE2 A0A0M5IZH3 A0A182XU94 A0A1W4UT22 A0A182II24 A0A3B0KEL4 B4KAI7 W8AZH0 A0A0Q9X4G1 A0A0Q9WPR8 A0A2S2QDI2 H9J5G2 B3LZ81 A0A2A4JDY9 Q7PUJ2 A0A026WSM2 A0A3L8D7Y1 A0A158NT60 A0A154P3M9 A0A151IA00 A0A2J7PYK2 A0A212FI98 A0A182UG36 A0A194RK28 A0A437BXX4 A0A194PPB3 A0A2H1WJ89 A0A151WJA9 A0A195EGJ5 A0A2J7PYN3 A0A0P9ANK1 A0A0K8T359 A0A0A9ZBH5 C0PTV4 B0WA44 A0A067RCL4 A0A1B6G259 A0A2A3E5P9 A0A182N5E5 E0VML7 A0A182R5H8 A0A088A033 A0A212EM07 A0A1J1HQ06 A0A2R7WZ49 B3LX91 A0A1Y1NKS5 A0A1W4UR97
Pubmed
EMBL
AGBW02008062
OWR54211.1
KQ459439
KPJ01023.1
KQ460397
KPJ15325.1
+ More
KZ150099 PZC73538.1 RSAL01000023 RVE52251.1 RVE52253.1 KPJ01021.1 KPJ15324.1 JTDY01003187 KOB69998.1 ODYU01009627 SOQ54194.1 AAZX01000985 AAZX01008934 GEZM01008094 JAV94862.1 JTDY01004822 KOB67707.1 KK107061 EZA61348.1 KQ435007 KZC13410.1 NNAY01000395 OXU28681.1 QOIP01000014 RLU14968.1 KQ981727 KYN37161.1 KQ435710 KOX79819.1 KQ979236 KYN22176.1 ADTU01011685 ADTU01011686 ADTU01011687 ADTU01011688 ADTU01011689 ADTU01011690 ADTU01011691 ADTU01011692 ADTU01011693 ADTU01011694 KQ976642 KYM78678.1 GBYB01006328 JAG76095.1 GBYB01000377 JAG70144.1 GEDC01008360 GEDC01006854 JAS28938.1 JAS30444.1 GBYB01000378 GBYB01001375 JAG70145.1 JAG71142.1 GDHC01005991 JAQ12638.1 GBHO01019822 JAG23782.1 GGMR01005320 MBY17939.1 GFXV01000223 MBW12028.1 CH477211 EAT47681.1 BT099830 ACV91668.1 KQ971354 EFA06209.2 KQ414588 KOC70468.1 ABLF02039633 ABLF02039634 ABLF02039636 ABLF02039639 ABLF02039640 ABLF02039642 ABLF02039645 ABLF02039646 ABLF02039647 ABLF02039648 AE014297 ACZ94883.1 KZ288301 PBC28857.1 BT030418 ABO52838.1 GBHO01004899 GBRD01005769 GDHC01012855 JAG38705.1 JAG60052.1 JAQ05774.1 UFQS01000043 UFQT01000043 SSW98369.1 SSX18755.1 AAAB01008987 DS235315 EEB14615.1 ATLV01015761 ATLV01015762 KE525036 KFB40641.1 ABI31162.1 CH480815 EDW42669.1 CM000160 EDW96867.2 CH954181 EDV49573.1 CH479185 EDW38366.1 CP012526 ALC45674.1 APCN01002092 APCN01002093 OUUW01000007 SPP83491.1 CH933806 EDW15700.2 GAMC01016297 GAMC01016296 GAMC01016295 JAB90258.1 CH964272 KRF99985.1 CH940650 KRF83023.1 GGMS01006538 MBY75741.1 BABH01028139 CH902617 EDV43008.2 NWSH01001734 PCG70297.1 EAA01423.6 KK107111 EZA59032.1 QOIP01000011 RLU16384.1 ADTU01025502 ADTU01025503 ADTU01025504 ADTU01025505 ADTU01025506 ADTU01025507 ADTU01025508 ADTU01025509 ADTU01025510 ADTU01025511 KQ434809 KZC06457.1 KQ978252 KYM95902.1 NEVH01020344 PNF21423.1 AGBW02008416 OWR53449.1 KQ460118 KPJ17690.1 RSAL01000001 RVE55298.1 KQ459596 KPI95281.1 ODYU01008707 SOQ52514.1 KQ983039 KYQ47942.1 KQ978957 KYN27022.1 PNF21422.1 KPU79298.1 GBRD01006221 JAG59600.1 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 BT071799 ACN43735.2 DS231868 EDS40777.1 KK852551 KDR21492.1 GECZ01013250 JAS56519.1 KZ288379 PBC26576.1 EEB14623.1 AGBW02013965 OWR42507.1 CVRI01000006 CRK88297.1 KK856079 PTY24802.1 EDV41691.1 KPU79296.1 KPU79297.1 KPU79299.1 GEZM01002471 JAV97235.1
KZ150099 PZC73538.1 RSAL01000023 RVE52251.1 RVE52253.1 KPJ01021.1 KPJ15324.1 JTDY01003187 KOB69998.1 ODYU01009627 SOQ54194.1 AAZX01000985 AAZX01008934 GEZM01008094 JAV94862.1 JTDY01004822 KOB67707.1 KK107061 EZA61348.1 KQ435007 KZC13410.1 NNAY01000395 OXU28681.1 QOIP01000014 RLU14968.1 KQ981727 KYN37161.1 KQ435710 KOX79819.1 KQ979236 KYN22176.1 ADTU01011685 ADTU01011686 ADTU01011687 ADTU01011688 ADTU01011689 ADTU01011690 ADTU01011691 ADTU01011692 ADTU01011693 ADTU01011694 KQ976642 KYM78678.1 GBYB01006328 JAG76095.1 GBYB01000377 JAG70144.1 GEDC01008360 GEDC01006854 JAS28938.1 JAS30444.1 GBYB01000378 GBYB01001375 JAG70145.1 JAG71142.1 GDHC01005991 JAQ12638.1 GBHO01019822 JAG23782.1 GGMR01005320 MBY17939.1 GFXV01000223 MBW12028.1 CH477211 EAT47681.1 BT099830 ACV91668.1 KQ971354 EFA06209.2 KQ414588 KOC70468.1 ABLF02039633 ABLF02039634 ABLF02039636 ABLF02039639 ABLF02039640 ABLF02039642 ABLF02039645 ABLF02039646 ABLF02039647 ABLF02039648 AE014297 ACZ94883.1 KZ288301 PBC28857.1 BT030418 ABO52838.1 GBHO01004899 GBRD01005769 GDHC01012855 JAG38705.1 JAG60052.1 JAQ05774.1 UFQS01000043 UFQT01000043 SSW98369.1 SSX18755.1 AAAB01008987 DS235315 EEB14615.1 ATLV01015761 ATLV01015762 KE525036 KFB40641.1 ABI31162.1 CH480815 EDW42669.1 CM000160 EDW96867.2 CH954181 EDV49573.1 CH479185 EDW38366.1 CP012526 ALC45674.1 APCN01002092 APCN01002093 OUUW01000007 SPP83491.1 CH933806 EDW15700.2 GAMC01016297 GAMC01016296 GAMC01016295 JAB90258.1 CH964272 KRF99985.1 CH940650 KRF83023.1 GGMS01006538 MBY75741.1 BABH01028139 CH902617 EDV43008.2 NWSH01001734 PCG70297.1 EAA01423.6 KK107111 EZA59032.1 QOIP01000011 RLU16384.1 ADTU01025502 ADTU01025503 ADTU01025504 ADTU01025505 ADTU01025506 ADTU01025507 ADTU01025508 ADTU01025509 ADTU01025510 ADTU01025511 KQ434809 KZC06457.1 KQ978252 KYM95902.1 NEVH01020344 PNF21423.1 AGBW02008416 OWR53449.1 KQ460118 KPJ17690.1 RSAL01000001 RVE55298.1 KQ459596 KPI95281.1 ODYU01008707 SOQ52514.1 KQ983039 KYQ47942.1 KQ978957 KYN27022.1 PNF21422.1 KPU79298.1 GBRD01006221 JAG59600.1 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 BT071799 ACN43735.2 DS231868 EDS40777.1 KK852551 KDR21492.1 GECZ01013250 JAS56519.1 KZ288379 PBC26576.1 EEB14623.1 AGBW02013965 OWR42507.1 CVRI01000006 CRK88297.1 KK856079 PTY24802.1 EDV41691.1 KPU79296.1 KPU79297.1 KPU79299.1 GEZM01002471 JAV97235.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000283053
UP000037510
UP000002358
+ More
UP000053097 UP000076502 UP000215335 UP000279307 UP000078541 UP000053105 UP000078492 UP000005205 UP000078540 UP000008820 UP000005203 UP000079169 UP000007266 UP000053825 UP000007819 UP000095300 UP000000803 UP000242457 UP000009046 UP000030765 UP000001292 UP000002282 UP000008711 UP000008744 UP000092553 UP000076407 UP000192221 UP000075840 UP000268350 UP000009192 UP000007798 UP000008792 UP000005204 UP000007801 UP000218220 UP000007062 UP000078542 UP000235965 UP000075902 UP000075809 UP000002320 UP000027135 UP000075884 UP000075900 UP000183832
UP000053097 UP000076502 UP000215335 UP000279307 UP000078541 UP000053105 UP000078492 UP000005205 UP000078540 UP000008820 UP000005203 UP000079169 UP000007266 UP000053825 UP000007819 UP000095300 UP000000803 UP000242457 UP000009046 UP000030765 UP000001292 UP000002282 UP000008711 UP000008744 UP000092553 UP000076407 UP000192221 UP000075840 UP000268350 UP000009192 UP000007798 UP000008792 UP000005204 UP000007801 UP000218220 UP000007062 UP000078542 UP000235965 UP000075902 UP000075809 UP000002320 UP000027135 UP000075884 UP000075900 UP000183832
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A212FKE7
A0A194Q659
A0A194RDG7
A0A2W1BFG7
A0A3S2P4N9
A0A437BPB1
+ More
A0A194QC59 A0A194RCY1 A0A0L7L3B9 A0A2H1WP36 K7J3W1 A0A1Y1NAB6 A0A0L7KWU5 A0A026WZS6 A0A154PQL6 A0A232FD16 A0A3L8D540 A0A195F9V8 A0A0M9ABL7 A0A195EAN5 A0A158NCS0 A0A151HZS0 A0A0C9PXU9 A0A0C9QIG5 A0A1B6DTE2 A0A0C9R081 A0A146LYE1 A0A0A9XSS5 A0A2S2NL76 A0A2H8TFI4 Q17LV3 C8VV84 A0A088AEQ8 A0A1S3D0I8 D6WPK0 A0A0L7RI46 J9K2X1 A0A1I8Q7E3 E1JII4 A0A2A3EAV1 A4IJ60 A0A0A9ZA85 A0A336LM08 A0A1S4GDS9 E0VMK9 A0A084VRP7 Q0KI85 B4HIF4 B4PLG0 B3NZK0 B4GLE2 A0A0M5IZH3 A0A182XU94 A0A1W4UT22 A0A182II24 A0A3B0KEL4 B4KAI7 W8AZH0 A0A0Q9X4G1 A0A0Q9WPR8 A0A2S2QDI2 H9J5G2 B3LZ81 A0A2A4JDY9 Q7PUJ2 A0A026WSM2 A0A3L8D7Y1 A0A158NT60 A0A154P3M9 A0A151IA00 A0A2J7PYK2 A0A212FI98 A0A182UG36 A0A194RK28 A0A437BXX4 A0A194PPB3 A0A2H1WJ89 A0A151WJA9 A0A195EGJ5 A0A2J7PYN3 A0A0P9ANK1 A0A0K8T359 A0A0A9ZBH5 C0PTV4 B0WA44 A0A067RCL4 A0A1B6G259 A0A2A3E5P9 A0A182N5E5 E0VML7 A0A182R5H8 A0A088A033 A0A212EM07 A0A1J1HQ06 A0A2R7WZ49 B3LX91 A0A1Y1NKS5 A0A1W4UR97
A0A194QC59 A0A194RCY1 A0A0L7L3B9 A0A2H1WP36 K7J3W1 A0A1Y1NAB6 A0A0L7KWU5 A0A026WZS6 A0A154PQL6 A0A232FD16 A0A3L8D540 A0A195F9V8 A0A0M9ABL7 A0A195EAN5 A0A158NCS0 A0A151HZS0 A0A0C9PXU9 A0A0C9QIG5 A0A1B6DTE2 A0A0C9R081 A0A146LYE1 A0A0A9XSS5 A0A2S2NL76 A0A2H8TFI4 Q17LV3 C8VV84 A0A088AEQ8 A0A1S3D0I8 D6WPK0 A0A0L7RI46 J9K2X1 A0A1I8Q7E3 E1JII4 A0A2A3EAV1 A4IJ60 A0A0A9ZA85 A0A336LM08 A0A1S4GDS9 E0VMK9 A0A084VRP7 Q0KI85 B4HIF4 B4PLG0 B3NZK0 B4GLE2 A0A0M5IZH3 A0A182XU94 A0A1W4UT22 A0A182II24 A0A3B0KEL4 B4KAI7 W8AZH0 A0A0Q9X4G1 A0A0Q9WPR8 A0A2S2QDI2 H9J5G2 B3LZ81 A0A2A4JDY9 Q7PUJ2 A0A026WSM2 A0A3L8D7Y1 A0A158NT60 A0A154P3M9 A0A151IA00 A0A2J7PYK2 A0A212FI98 A0A182UG36 A0A194RK28 A0A437BXX4 A0A194PPB3 A0A2H1WJ89 A0A151WJA9 A0A195EGJ5 A0A2J7PYN3 A0A0P9ANK1 A0A0K8T359 A0A0A9ZBH5 C0PTV4 B0WA44 A0A067RCL4 A0A1B6G259 A0A2A3E5P9 A0A182N5E5 E0VML7 A0A182R5H8 A0A088A033 A0A212EM07 A0A1J1HQ06 A0A2R7WZ49 B3LX91 A0A1Y1NKS5 A0A1W4UR97
PDB
3DMK
E-value=1.45741e-10,
Score=159
Ontologies
GO
Topology
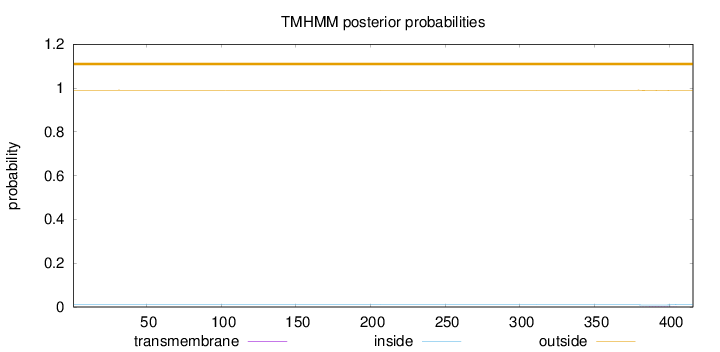
Length:
416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02743
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.00976
outside
1 - 416
Population Genetic Test Statistics
Pi
25.047317
Theta
19.792212
Tajima's D
0.759785
CLR
0.790309
CSRT
0.6015199240038
Interpretation
Uncertain
