Pre Gene Modal
BGIBMGA001115
Annotation
PREDICTED:_dolichyl-phosphate_beta-glucosyltransferase_[Papilio_polytes]
Full name
Dolichyl-phosphate beta-glucosyltransferase
Alternative Name
Wollknaeuel
Location in the cell
PlasmaMembrane Reliability : 3.672
Sequence
CDS
ATGGATCTCTTAACAATATTTCTGGTAATGGTTTTATATGGTATTGCTATAGCGTTGGCTCTATTAATTGTGCTCGTTATAATACTATATATAATAACCGATACGTATCCTGTTGTGGAGCGGTATAAAGATGAAGAAACATACAATGATTACTTAACAAATACTAAACTGAGATTCCCCTCTATACTTGAAGAAGAATCTGTTAGCTTAAGTGTAGTTGTTCCTGCATATAATGAAGAAAAAAGACTGCCACCCATGTTGGATGAAACAATTGAGTTCTTGGAGAACCGACAAAAAGAAAACCCTTCATACAAATATGAAATAATCATAGTGAGTGACGGAAGTAAAGACAGCACAGTTAAAGTAGCCGAGAGCTACTCCATAAAATATGGTAGTGACAAAGTGAAATGCCTAGAGTTAATAAAGAACAGAGGAAAAGGAGGAGCAGTCAGACTGGGCATACAAAGTTCGAGAGGTGCAACGATATTGTTTGCAGATGCAGATGGTGCCTCAAAATTTGAAGATTTGACGAAATTAGAAGTCGCCCTAAAGGATATTGTGAAATGTGATCCCCTCAAAGATGTTAAAACAACTAGTGAAAGTCTAGGAATTGTAATTGGTTCGAGGGCTCATCTTGAAAAGGAATCTTTAGCAAAAAGGAATATTTTTAGGAATATACTTATGTATGGATTTCATTTTCTGGTCTGGTTGTTCACTGTAAAAGGTATAAAAGACACACAGTGTGGCTTCAAACTTTTCACTAGAAAAGCAGCAAGAATATGTTTTGAAAGCTTACATGTAAACAGATGGGCATTTGATGTAGAGTTATTGTATATCGCACAAAAATTAAACATTCCAATATCCGAAATACCAGTGAGGTGGACAGAAATCGAAGGATCAAAAGTCACGCCAGTTGTCGCTTGGATACAAATGGGATGTGACCTAGGCCTTATCTGGCTTAAATATAGAATAGGAGCTTGGAAGATAAAAAGCGAAAAAACAGAGTGA
Protein
MDLLTIFLVMVLYGIAIALALLIVLVIILYIITDTYPVVERYKDEETYNDYLTNTKLRFPSILEEESVSLSVVVPAYNEEKRLPPMLDETIEFLENRQKENPSYKYEIIIVSDGSKDSTVKVAESYSIKYGSDKVKCLELIKNRGKGGAVRLGIQSSRGATILFADADGASKFEDLTKLEVALKDIVKCDPLKDVKTTSESLGIVIGSRAHLEKESLAKRNIFRNILMYGFHFLVWLFTVKGIKDTQCGFKLFTRKAARICFESLHVNRWAFDVELLYIAQKLNIPISEIPVRWTEIEGSKVTPVVAWIQMGCDLGLIWLKYRIGAWKIKSEKTE
Summary
Description
Required for normal production of N-glycosylated proteins in the endoplasmic reticulum (ER). Required for embryonic segmentation, dorsal-ventral patterning and gastrulation. Required for chitin orientation and shaping of the apical and lateral plasma membranes of epidermal cells during cuticle differentiation. Also required for correctly shaping apical membrane topology of the epithelia of other organs such as the midgut and the hindgut.
Catalytic Activity
dolichyl phosphate + UDP-alpha-D-glucose = dolichyl beta-D-glucosyl phosphate + UDP
Miscellaneous
'Wollknaeuel' means 'ball of wool' in German.
Similarity
Belongs to the glycosyltransferase 2 family.
Keywords
Complete proteome
Endoplasmic reticulum
Glycosyltransferase
Membrane
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Dolichyl-phosphate beta-glucosyltransferase
Uniprot
H9IV35
I4DJP6
A0A194Q6L7
A0A2A4JCR7
A0A2W1BXM9
A0A3S2LRI2
+ More
A0A212ERS4 S4PWG3 A0A2H1W5E8 J3JX27 R4WPG9 D2A2D5 A0A1B6E2H3 A0A1W4WHF5 N6TX99 E0VH07 A0A1B6MTM3 A0A224XTH8 A0A067R8A0 A0A146MBZ4 A0A0A9WIG3 A0A2J7R8T5 A0A2P2HW56 A0A2R7VZ92 Q1HQV4 T1HHV7 A0A226E7B7 A0A1W4V7F2 B4Q6C9 B3N733 Q9VLQ1 B3MPG9 B4HYH8 B4NWV4 A0A023EPS4 A0A1Q3FAZ5 E9IJH2 A0A0P5LD01 A0A1Y1K4B3 A0A0P5JZP3 A0A2R5LKZ9 A0A0P5YGC1 A0A0C9RMI0 A0A0P6DYT1 A0A151J999 A0A0P5ZV63 A0A151IM96 A0A0P6EFT9 A0A293MSJ0 A0A131XEG2 A0A224YXL7 A0A131Z2U5 Q29MM8 A0A0P5VAT3 B4G906 A0A158NBT3 F4X8A4 A0A1A9Y3K1 A0A1B0BLA3 L7MIJ2 A0A1Z5KYY3 A0A3P9DFF1 A0A182Y3D5 A0A3B0JBS7 B4KJM4 A0A026W2S2 A0A3Q2VD11 A0A3B4G6E0 A0A0P5TBW9 A0A084WBV6 A0A3P8NID8 A0A195FDB8 M4A2D9 A0A3Q2WIJ1 A0A182SY43 A0A182JYM0 A0A151X9I1 A0A3P9DFD2 A0A3P9P5N1 A0A0P5TLA1 B4LSH2 A0A3B4UM81 A0A3B4YP45 A0A3Q4HIH6 A0A3P8NII2 A0A1B0AEN9 D3TQ93 A0A2U9B7V8 B4MTJ6 A0A3Q2Q6G8 E9FRI2 A0A0P5Q8P3 A0A195BG85 A0A1A9WWC1 A0A0E9X8S6 A0A315VIV5 A0A232F742 I3K425 A0A3B3UVA6 A0A154NXS1
A0A212ERS4 S4PWG3 A0A2H1W5E8 J3JX27 R4WPG9 D2A2D5 A0A1B6E2H3 A0A1W4WHF5 N6TX99 E0VH07 A0A1B6MTM3 A0A224XTH8 A0A067R8A0 A0A146MBZ4 A0A0A9WIG3 A0A2J7R8T5 A0A2P2HW56 A0A2R7VZ92 Q1HQV4 T1HHV7 A0A226E7B7 A0A1W4V7F2 B4Q6C9 B3N733 Q9VLQ1 B3MPG9 B4HYH8 B4NWV4 A0A023EPS4 A0A1Q3FAZ5 E9IJH2 A0A0P5LD01 A0A1Y1K4B3 A0A0P5JZP3 A0A2R5LKZ9 A0A0P5YGC1 A0A0C9RMI0 A0A0P6DYT1 A0A151J999 A0A0P5ZV63 A0A151IM96 A0A0P6EFT9 A0A293MSJ0 A0A131XEG2 A0A224YXL7 A0A131Z2U5 Q29MM8 A0A0P5VAT3 B4G906 A0A158NBT3 F4X8A4 A0A1A9Y3K1 A0A1B0BLA3 L7MIJ2 A0A1Z5KYY3 A0A3P9DFF1 A0A182Y3D5 A0A3B0JBS7 B4KJM4 A0A026W2S2 A0A3Q2VD11 A0A3B4G6E0 A0A0P5TBW9 A0A084WBV6 A0A3P8NID8 A0A195FDB8 M4A2D9 A0A3Q2WIJ1 A0A182SY43 A0A182JYM0 A0A151X9I1 A0A3P9DFD2 A0A3P9P5N1 A0A0P5TLA1 B4LSH2 A0A3B4UM81 A0A3B4YP45 A0A3Q4HIH6 A0A3P8NII2 A0A1B0AEN9 D3TQ93 A0A2U9B7V8 B4MTJ6 A0A3Q2Q6G8 E9FRI2 A0A0P5Q8P3 A0A195BG85 A0A1A9WWC1 A0A0E9X8S6 A0A315VIV5 A0A232F742 I3K425 A0A3B3UVA6 A0A154NXS1
EC Number
2.4.1.117
Pubmed
19121390
22651552
26354079
28756777
22118469
23622113
+ More
22516182 23691247 18362917 19820115 23537049 20566863 24845553 26823975 25401762 17204158 17510324 17994087 22936249 10731132 12537572 12537569 18403407 21199819 17550304 24945155 21282665 28004739 28049606 28797301 26830274 15632085 21347285 21719571 25576852 28528879 25186727 25244985 24508170 30249741 24438588 23542700 20353571 21292972 25613341 29703783 28648823
22516182 23691247 18362917 19820115 23537049 20566863 24845553 26823975 25401762 17204158 17510324 17994087 22936249 10731132 12537572 12537569 18403407 21199819 17550304 24945155 21282665 28004739 28049606 28797301 26830274 15632085 21347285 21719571 25576852 28528879 25186727 25244985 24508170 30249741 24438588 23542700 20353571 21292972 25613341 29703783 28648823
EMBL
BABH01000380
AK401514
BAM18136.1
KQ459439
KPJ01019.1
NWSH01001864
+ More
PCG69875.1 KZ149891 PZC79081.1 RSAL01000023 RVE52274.1 AGBW02012947 OWR44187.1 GAIX01006838 JAA85722.1 ODYU01006452 SOQ48310.1 BT127795 AEE62757.1 AK417551 BAN20766.1 KQ971338 EFA02177.1 GEDC01022662 GEDC01005172 JAS14636.1 JAS32126.1 APGK01010787 KB738301 KB632179 ENN82673.1 ERL89607.1 DS235156 EEB12663.1 GEBQ01000732 JAT39245.1 GFTR01005147 JAW11279.1 KK852637 KDR19680.1 GDHC01001331 JAQ17298.1 GBHO01038984 GBRD01015182 JAG04620.1 JAG50644.1 NEVH01006721 PNF37240.1 IACF01000178 LAB65982.1 KK854141 PTY12000.1 DQ440340 CH477261 ABF18373.1 EAT45653.1 ACPB03018544 LNIX01000006 OXA53248.1 CM000361 CM002910 EDX04205.1 KMY89030.1 CH954177 EDV59329.1 AE014134 AY094692 AAF52633.1 AAM11045.1 CH902620 EDV31265.1 CH480818 EDW52108.1 CM000157 EDW88484.1 GAPW01002555 JAC11043.1 GFDL01010304 JAV24741.1 GL763802 EFZ19267.1 GDIQ01178928 GDIP01090166 GDIP01087718 GDIP01011833 GDIQ01084258 GDIQ01064775 JAK72797.1 JAM13549.1 GEZM01095847 JAV55298.1 GDIQ01191868 JAK59857.1 GGLE01006046 MBY10172.1 GDIP01168455 GDIP01164650 GDIQ01228113 GDIQ01223565 GDIQ01125904 GDIQ01091147 GDIP01090165 GDIP01088817 GDIP01083764 GDIP01072064 JAK23612.1 JAM31651.1 GBYB01008186 GBYB01008190 JAG77953.1 JAG77957.1 GDIQ01072069 JAN22668.1 KQ979433 KYN21591.1 GDIP01232590 GDIQ01145446 GDIP01137409 GDIP01098462 GDIP01073313 GDIP01039009 JAL06280.1 JAM64706.1 KQ977059 KYN06012.1 GDIP01232589 GDIP01192300 GDIP01137408 GDIP01123151 GDIP01098463 GDIP01073314 GDIQ01077562 JAI90812.1 JAN17175.1 GFWV01019076 MAA43804.1 GEFH01004121 JAP64460.1 GFPF01007374 MAA18520.1 GEDV01002824 JAP85733.1 CH379060 EAL33665.2 GDIP01102955 JAM00760.1 CH479180 EDW28836.1 ADTU01011316 GL888932 EGI57178.1 JXJN01016270 GACK01001003 JAA64031.1 GFJQ02006969 JAW00001.1 OUUW01000004 SPP79804.1 CH933807 EDW11469.1 KK107459 QOIP01000004 EZA50377.1 RLU23708.1 GDIP01128250 JAL75464.1 ATLV01022472 KE525332 KFB47700.1 KQ981673 KYN38376.1 KQ982373 KYQ57023.1 GDIP01128208 JAL75506.1 CH940649 EDW64794.1 EZ423595 ADD19871.1 CP026246 AWO99915.1 AWO99916.1 CH963852 EDW75435.1 GL732523 EFX90194.1 GDIQ01117652 JAL34074.1 KQ976500 KYM83175.1 GBXM01010347 JAH98230.1 NHOQ01001678 PWA22876.1 NNAY01000752 OXU26676.1 AERX01050045 KQ434778 KZC04417.1
PCG69875.1 KZ149891 PZC79081.1 RSAL01000023 RVE52274.1 AGBW02012947 OWR44187.1 GAIX01006838 JAA85722.1 ODYU01006452 SOQ48310.1 BT127795 AEE62757.1 AK417551 BAN20766.1 KQ971338 EFA02177.1 GEDC01022662 GEDC01005172 JAS14636.1 JAS32126.1 APGK01010787 KB738301 KB632179 ENN82673.1 ERL89607.1 DS235156 EEB12663.1 GEBQ01000732 JAT39245.1 GFTR01005147 JAW11279.1 KK852637 KDR19680.1 GDHC01001331 JAQ17298.1 GBHO01038984 GBRD01015182 JAG04620.1 JAG50644.1 NEVH01006721 PNF37240.1 IACF01000178 LAB65982.1 KK854141 PTY12000.1 DQ440340 CH477261 ABF18373.1 EAT45653.1 ACPB03018544 LNIX01000006 OXA53248.1 CM000361 CM002910 EDX04205.1 KMY89030.1 CH954177 EDV59329.1 AE014134 AY094692 AAF52633.1 AAM11045.1 CH902620 EDV31265.1 CH480818 EDW52108.1 CM000157 EDW88484.1 GAPW01002555 JAC11043.1 GFDL01010304 JAV24741.1 GL763802 EFZ19267.1 GDIQ01178928 GDIP01090166 GDIP01087718 GDIP01011833 GDIQ01084258 GDIQ01064775 JAK72797.1 JAM13549.1 GEZM01095847 JAV55298.1 GDIQ01191868 JAK59857.1 GGLE01006046 MBY10172.1 GDIP01168455 GDIP01164650 GDIQ01228113 GDIQ01223565 GDIQ01125904 GDIQ01091147 GDIP01090165 GDIP01088817 GDIP01083764 GDIP01072064 JAK23612.1 JAM31651.1 GBYB01008186 GBYB01008190 JAG77953.1 JAG77957.1 GDIQ01072069 JAN22668.1 KQ979433 KYN21591.1 GDIP01232590 GDIQ01145446 GDIP01137409 GDIP01098462 GDIP01073313 GDIP01039009 JAL06280.1 JAM64706.1 KQ977059 KYN06012.1 GDIP01232589 GDIP01192300 GDIP01137408 GDIP01123151 GDIP01098463 GDIP01073314 GDIQ01077562 JAI90812.1 JAN17175.1 GFWV01019076 MAA43804.1 GEFH01004121 JAP64460.1 GFPF01007374 MAA18520.1 GEDV01002824 JAP85733.1 CH379060 EAL33665.2 GDIP01102955 JAM00760.1 CH479180 EDW28836.1 ADTU01011316 GL888932 EGI57178.1 JXJN01016270 GACK01001003 JAA64031.1 GFJQ02006969 JAW00001.1 OUUW01000004 SPP79804.1 CH933807 EDW11469.1 KK107459 QOIP01000004 EZA50377.1 RLU23708.1 GDIP01128250 JAL75464.1 ATLV01022472 KE525332 KFB47700.1 KQ981673 KYN38376.1 KQ982373 KYQ57023.1 GDIP01128208 JAL75506.1 CH940649 EDW64794.1 EZ423595 ADD19871.1 CP026246 AWO99915.1 AWO99916.1 CH963852 EDW75435.1 GL732523 EFX90194.1 GDIQ01117652 JAL34074.1 KQ976500 KYM83175.1 GBXM01010347 JAH98230.1 NHOQ01001678 PWA22876.1 NNAY01000752 OXU26676.1 AERX01050045 KQ434778 KZC04417.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000283053
UP000007151
UP000007266
+ More
UP000192223 UP000019118 UP000030742 UP000009046 UP000027135 UP000235965 UP000008820 UP000015103 UP000198287 UP000192221 UP000000304 UP000008711 UP000000803 UP000007801 UP000001292 UP000002282 UP000078492 UP000078542 UP000001819 UP000008744 UP000005205 UP000007755 UP000092443 UP000092460 UP000265160 UP000076408 UP000268350 UP000009192 UP000053097 UP000279307 UP000264840 UP000261460 UP000030765 UP000265100 UP000078541 UP000002852 UP000075901 UP000075881 UP000075809 UP000242638 UP000008792 UP000261420 UP000261360 UP000261580 UP000092445 UP000246464 UP000007798 UP000265000 UP000000305 UP000078540 UP000091820 UP000215335 UP000005207 UP000261500 UP000076502
UP000192223 UP000019118 UP000030742 UP000009046 UP000027135 UP000235965 UP000008820 UP000015103 UP000198287 UP000192221 UP000000304 UP000008711 UP000000803 UP000007801 UP000001292 UP000002282 UP000078492 UP000078542 UP000001819 UP000008744 UP000005205 UP000007755 UP000092443 UP000092460 UP000265160 UP000076408 UP000268350 UP000009192 UP000053097 UP000279307 UP000264840 UP000261460 UP000030765 UP000265100 UP000078541 UP000002852 UP000075901 UP000075881 UP000075809 UP000242638 UP000008792 UP000261420 UP000261360 UP000261580 UP000092445 UP000246464 UP000007798 UP000265000 UP000000305 UP000078540 UP000091820 UP000215335 UP000005207 UP000261500 UP000076502
Interpro
IPR029044
Nucleotide-diphossugar_trans
+ More
IPR035518 DPG_synthase
IPR001173 Glyco_trans_2-like
IPR009030 Growth_fac_rcpt_cys_sf
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR001881 EGF-like_Ca-bd_dom
IPR000742 EGF-like_dom
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR035518 DPG_synthase
IPR001173 Glyco_trans_2-like
IPR009030 Growth_fac_rcpt_cys_sf
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR001881 EGF-like_Ca-bd_dom
IPR000742 EGF-like_dom
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
Gene 3D
ProteinModelPortal
H9IV35
I4DJP6
A0A194Q6L7
A0A2A4JCR7
A0A2W1BXM9
A0A3S2LRI2
+ More
A0A212ERS4 S4PWG3 A0A2H1W5E8 J3JX27 R4WPG9 D2A2D5 A0A1B6E2H3 A0A1W4WHF5 N6TX99 E0VH07 A0A1B6MTM3 A0A224XTH8 A0A067R8A0 A0A146MBZ4 A0A0A9WIG3 A0A2J7R8T5 A0A2P2HW56 A0A2R7VZ92 Q1HQV4 T1HHV7 A0A226E7B7 A0A1W4V7F2 B4Q6C9 B3N733 Q9VLQ1 B3MPG9 B4HYH8 B4NWV4 A0A023EPS4 A0A1Q3FAZ5 E9IJH2 A0A0P5LD01 A0A1Y1K4B3 A0A0P5JZP3 A0A2R5LKZ9 A0A0P5YGC1 A0A0C9RMI0 A0A0P6DYT1 A0A151J999 A0A0P5ZV63 A0A151IM96 A0A0P6EFT9 A0A293MSJ0 A0A131XEG2 A0A224YXL7 A0A131Z2U5 Q29MM8 A0A0P5VAT3 B4G906 A0A158NBT3 F4X8A4 A0A1A9Y3K1 A0A1B0BLA3 L7MIJ2 A0A1Z5KYY3 A0A3P9DFF1 A0A182Y3D5 A0A3B0JBS7 B4KJM4 A0A026W2S2 A0A3Q2VD11 A0A3B4G6E0 A0A0P5TBW9 A0A084WBV6 A0A3P8NID8 A0A195FDB8 M4A2D9 A0A3Q2WIJ1 A0A182SY43 A0A182JYM0 A0A151X9I1 A0A3P9DFD2 A0A3P9P5N1 A0A0P5TLA1 B4LSH2 A0A3B4UM81 A0A3B4YP45 A0A3Q4HIH6 A0A3P8NII2 A0A1B0AEN9 D3TQ93 A0A2U9B7V8 B4MTJ6 A0A3Q2Q6G8 E9FRI2 A0A0P5Q8P3 A0A195BG85 A0A1A9WWC1 A0A0E9X8S6 A0A315VIV5 A0A232F742 I3K425 A0A3B3UVA6 A0A154NXS1
A0A212ERS4 S4PWG3 A0A2H1W5E8 J3JX27 R4WPG9 D2A2D5 A0A1B6E2H3 A0A1W4WHF5 N6TX99 E0VH07 A0A1B6MTM3 A0A224XTH8 A0A067R8A0 A0A146MBZ4 A0A0A9WIG3 A0A2J7R8T5 A0A2P2HW56 A0A2R7VZ92 Q1HQV4 T1HHV7 A0A226E7B7 A0A1W4V7F2 B4Q6C9 B3N733 Q9VLQ1 B3MPG9 B4HYH8 B4NWV4 A0A023EPS4 A0A1Q3FAZ5 E9IJH2 A0A0P5LD01 A0A1Y1K4B3 A0A0P5JZP3 A0A2R5LKZ9 A0A0P5YGC1 A0A0C9RMI0 A0A0P6DYT1 A0A151J999 A0A0P5ZV63 A0A151IM96 A0A0P6EFT9 A0A293MSJ0 A0A131XEG2 A0A224YXL7 A0A131Z2U5 Q29MM8 A0A0P5VAT3 B4G906 A0A158NBT3 F4X8A4 A0A1A9Y3K1 A0A1B0BLA3 L7MIJ2 A0A1Z5KYY3 A0A3P9DFF1 A0A182Y3D5 A0A3B0JBS7 B4KJM4 A0A026W2S2 A0A3Q2VD11 A0A3B4G6E0 A0A0P5TBW9 A0A084WBV6 A0A3P8NID8 A0A195FDB8 M4A2D9 A0A3Q2WIJ1 A0A182SY43 A0A182JYM0 A0A151X9I1 A0A3P9DFD2 A0A3P9P5N1 A0A0P5TLA1 B4LSH2 A0A3B4UM81 A0A3B4YP45 A0A3Q4HIH6 A0A3P8NII2 A0A1B0AEN9 D3TQ93 A0A2U9B7V8 B4MTJ6 A0A3Q2Q6G8 E9FRI2 A0A0P5Q8P3 A0A195BG85 A0A1A9WWC1 A0A0E9X8S6 A0A315VIV5 A0A232F742 I3K425 A0A3B3UVA6 A0A154NXS1
PDB
5EKE
E-value=6.48646e-05,
Score=109
Ontologies
PATHWAY
GO
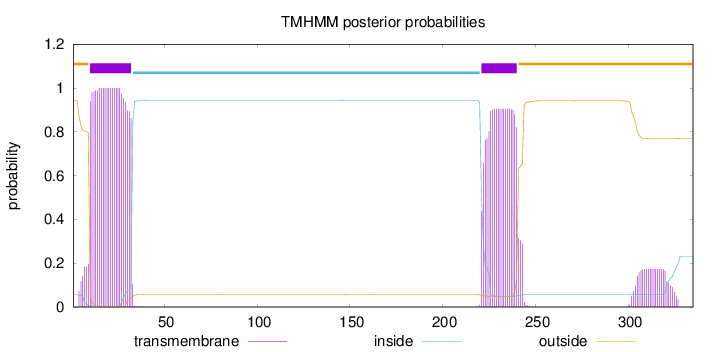
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
335
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.7803
Exp number, first 60 AAs:
23.42425
Total prob of N-in:
0.05651
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 220
TMhelix
221 - 240
outside
241 - 335
Population Genetic Test Statistics
Pi
21.772285
Theta
19.914223
Tajima's D
0.256311
CLR
4.510425
CSRT
0.455927203639818
Interpretation
Uncertain
