Gene
KWMTBOMO08061
Pre Gene Modal
BGIBMGA001118
Annotation
PREDICTED:_7-methylguanosine_phosphate-specific_5'-nucleotidase_[Papilio_machaon]
Full name
5'-nucleotidase
+ More
7-methylguanosine phosphate-specific 5'-nucleotidase
7-methylguanosine phosphate-specific 5'-nucleotidase
Alternative Name
Cytosolic 5'-nucleotidase IIIB
N(7)-methylguanylate 5'-phosphatase
N(7)-methylguanylate 5'-phosphatase
Location in the cell
Cytoplasmic Reliability : 1.333 Mitochondrial Reliability : 1.309 Nuclear Reliability : 1.384
Sequence
CDS
ATGAAGGTTTTAACGAGGCTTCAAGAAATTCCGGAGTTTAACAGAGAAAACATCCATATTAGAGACGAGACGGAACTAGTCGCGAAGTTAAATAGAATCATAAAAGGCGGACACAACAAATTGCAGATAGTGACAGATTTTGACCATACGCTGACAAGACACACTTTGGATAATGGAAAATCCGTTCTCACTAGTTTTGGTATGTGA
Protein
MKVLTRLQEIPEFNRENIHIRDETELVAKLNRIIKGGHNKLQIVTDFDHTLTRHTLDNGKSVLTSFGM
Summary
Description
Specifically hydrolyzes 7-methylguanosine monophosphate (m(7)GMP) to 7-methylguanosine and inorganic phosphate (PubMed:23223233, PubMed:24603684). Also able to mediate hydrolysis of diphosphate (m(7)GDP) to 7-methylguanosine and 2 inorganic phosphate with lower activity (PubMed:23223233). The specific activity for m(7)GMP may protect cells against undesired salvage of m(7)GMP and its incorporation into nucleic acids (PubMed:23223233). Also has weak activity for CMP (PubMed:23223233, PubMed:24603684). UMP and purine nucleotides are poor substrates (PubMed:23223233, PubMed:24603684).
Catalytic Activity
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
7-methyl-GMP + H2O = N(7)-methylguanosine + phosphate
CMP + H2O = cytidine + phosphate
7-methyl-GMP + H2O = N(7)-methylguanosine + phosphate
CMP + H2O = cytidine + phosphate
Cofactor
Mg(2+)
Biophysicochemical Properties
13 uM for m(7)GMP (at 25 degrees Celsius)
12 uM for m(7)GMP (at 25 degrees Celsius)
48 uM for CMP (at 25 degrees Celsius)
102 uM for GMP (at 25 degrees Celsius)
32 uM for AMP (at 25 degrees Celsius)
91 uM for UMP (at 25 degrees Celsius)
12 uM for m(7)GMP (at 25 degrees Celsius)
48 uM for CMP (at 25 degrees Celsius)
102 uM for GMP (at 25 degrees Celsius)
32 uM for AMP (at 25 degrees Celsius)
91 uM for UMP (at 25 degrees Celsius)
Subunit
Monomer.
Similarity
Belongs to the pyrimidine 5'-nucleotidase family.
Keywords
3D-structure
Complete proteome
Hydrolase
Magnesium
Metal-binding
Nucleotide metabolism
Nucleotide-binding
Reference proteome
Feature
chain 7-methylguanosine phosphate-specific 5'-nucleotidase
Uniprot
H9IV38
A0A2A4JCN7
A0A2W1C271
A0A194RDF8
A0A2H1VTT7
A0A3S2PIF6
+ More
A0A194Q6L2 S4PX53 A0A1E1WSJ8 A0A0L7KR62 A0A212ERR6 A0A182FVZ2 B0X0T4 A0A182JQ60 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 A0A0L7LM38 W5JP62 A0A182PN54 A0A336LZD2 A0A182J590 A0A084WUG0 A0A182KUZ0 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A0L7KRZ7 A0A2M4ALI7 T1E8J9 A0A182UBB7 A0A182NPA1 A0A182YAB5 A0A182Q2A3 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182RLV4 A0A1B0DDY6 Q17PQ5 A0A023EQ93 U5EYF0 A0A0M4EH73 B4KM33 A0A026W2Z4 E2A4U6 A0A034WB86 A0A1B0CBQ1 A0A1I8NLH3 A0A0J7KV79 W8BJC5 A0A2G9S4V8 E2C413 A0A0A1WQY6 Q9W197 A0A034W9L4 A0A0K8VG61 A0A195BG84 A0A195FDT2 A0A195DRX9 A0A158NBS5 B4I2F1 B4QBK0 A0A1B6KYL7 A0A1B6H3L6 A0A1B6KZG4 B3MDM8 A0A2P8Z7N4 A0A2G9S4X1 A0A0L0C605 A0A1Y1M1Z2 A0A1B6FFA8 A0A3N0YA27 A0A1A9ZQS9 A0A1A9UER1 A0A1B0FE50 A0A0T6B439 B4PAW2 A0A1W4UM40 A0A151X9L3 D1ZZV3 B4LK41 A0A023F065 A0A1L8DXW8 A0A232FCJ5 K7J061 A0A088ATS9 A0A0P4VK67 A0A2D4IWN5 A0A1L8DXY4 A0A2D4IWS6 A0A1B6KHD2 A0A1B6C974 B4J4S4 A0A1B6MFL6 S4RFV0 F7DEV4 D3TP59 A0A1B0BYQ4
A0A194Q6L2 S4PX53 A0A1E1WSJ8 A0A0L7KR62 A0A212ERR6 A0A182FVZ2 B0X0T4 A0A182JQ60 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 A0A0L7LM38 W5JP62 A0A182PN54 A0A336LZD2 A0A182J590 A0A084WUG0 A0A182KUZ0 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A0L7KRZ7 A0A2M4ALI7 T1E8J9 A0A182UBB7 A0A182NPA1 A0A182YAB5 A0A182Q2A3 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182RLV4 A0A1B0DDY6 Q17PQ5 A0A023EQ93 U5EYF0 A0A0M4EH73 B4KM33 A0A026W2Z4 E2A4U6 A0A034WB86 A0A1B0CBQ1 A0A1I8NLH3 A0A0J7KV79 W8BJC5 A0A2G9S4V8 E2C413 A0A0A1WQY6 Q9W197 A0A034W9L4 A0A0K8VG61 A0A195BG84 A0A195FDT2 A0A195DRX9 A0A158NBS5 B4I2F1 B4QBK0 A0A1B6KYL7 A0A1B6H3L6 A0A1B6KZG4 B3MDM8 A0A2P8Z7N4 A0A2G9S4X1 A0A0L0C605 A0A1Y1M1Z2 A0A1B6FFA8 A0A3N0YA27 A0A1A9ZQS9 A0A1A9UER1 A0A1B0FE50 A0A0T6B439 B4PAW2 A0A1W4UM40 A0A151X9L3 D1ZZV3 B4LK41 A0A023F065 A0A1L8DXW8 A0A232FCJ5 K7J061 A0A088ATS9 A0A0P4VK67 A0A2D4IWN5 A0A1L8DXY4 A0A2D4IWS6 A0A1B6KHD2 A0A1B6C974 B4J4S4 A0A1B6MFL6 S4RFV0 F7DEV4 D3TP59 A0A1B0BYQ4
EC Number
3.1.3.5
3.1.3.91
3.1.3.91
Pubmed
19121390
28756777
26354079
23622113
26227816
22118469
+ More
20920257 23761445 24438588 20966253 12364791 14747013 17210077 25244985 17510324 24945155 17994087 18057021 24508170 30249741 20798317 25348373 24495485 25830018 10731132 12537572 12537569 23223233 24603684 21347285 22936249 29403074 26108605 28004739 17550304 18362917 19820115 25474469 28648823 20075255 27129103 17495919 20353571
20920257 23761445 24438588 20966253 12364791 14747013 17210077 25244985 17510324 24945155 17994087 18057021 24508170 30249741 20798317 25348373 24495485 25830018 10731132 12537572 12537569 23223233 24603684 21347285 22936249 29403074 26108605 28004739 17550304 18362917 19820115 25474469 28648823 20075255 27129103 17495919 20353571
EMBL
BABH01000388
NWSH01001864
PCG69867.1
KZ149891
PZC79076.1
KQ460397
+ More
KPJ15315.1 ODYU01004396 SOQ44230.1 RSAL01000023 RVE52277.1 KQ459439 KPJ01014.1 GAIX01005638 JAA86922.1 GDQN01010614 GDQN01004724 GDQN01001049 JAT80440.1 JAT86330.1 JAT90005.1 JTDY01006698 KOB65763.1 AGBW02012947 OWR44182.1 DS232245 EDS38326.1 GFDL01008250 JAV26795.1 GGFM01007354 MBW28105.1 GGFJ01007807 MBW56948.1 JTDY01000627 KOB76419.1 ADMH02000766 ETN65108.1 UFQT01000339 SSX23354.1 ATLV01027083 KE525423 KFB53854.1 AAAB01008848 EAA07027.4 APCN01003547 JTDY01006610 KOB65836.1 GGFK01008332 MBW41653.1 GAMD01002708 JAA98882.1 AXCN02001814 AXCM01001000 AJVK01014661 CH477190 EAT48696.1 GAPW01002874 JAC10724.1 GANO01002042 JAB57829.1 CP012524 ALC41872.1 CH933808 EDW10822.1 KK107459 QOIP01000004 EZA50393.1 RLU24234.1 GL436748 EFN71540.1 GAKP01007552 JAC51400.1 AJWK01005748 AJWK01005749 LBMM01002768 KMQ94397.1 GAMC01007708 JAB98847.1 KV925179 PIO35124.1 GL452364 EFN77399.1 GBXI01013035 JAD01257.1 AE013599 AY061358 GAKP01007553 JAC51399.1 GDHF01014400 JAI37914.1 KQ976500 KYM83187.1 KQ981673 KYN38362.1 KQ980581 KYN15269.1 ADTU01011295 CH480820 EDW53946.1 CM000362 CM002911 EDX08518.1 KMY96290.1 GEBQ01023476 JAT16501.1 GECZ01000508 JAS69261.1 GEBQ01023192 JAT16785.1 CH902619 EDV37491.1 PYGN01000160 PSN52497.1 PIO35125.1 JRES01000857 KNC27715.1 GEZM01042433 JAV79854.1 GECZ01029236 GECZ01020891 GECZ01003524 JAS40533.1 JAS48878.1 JAS66245.1 RJVU01048900 ROL43072.1 CCAG010006200 LJIG01009893 KRT82170.1 CM000158 EDW92502.1 KQ982373 KYQ57009.1 KQ971338 EFA01792.1 CH940648 EDW60630.1 GBBI01003815 JAC14897.1 GFDF01002777 JAV11307.1 NNAY01000467 OXU28168.1 GDKW01001904 JAI54691.1 IACK01139303 LAA88665.1 GFDF01002776 JAV11308.1 IACK01139301 LAA88662.1 GEBQ01029413 JAT10564.1 GEDC01027265 GEDC01026000 JAS10033.1 JAS11298.1 CH916367 EDW00620.1 GEBQ01005298 JAT34679.1 EZ423211 ADD19487.1 JXJN01022766
KPJ15315.1 ODYU01004396 SOQ44230.1 RSAL01000023 RVE52277.1 KQ459439 KPJ01014.1 GAIX01005638 JAA86922.1 GDQN01010614 GDQN01004724 GDQN01001049 JAT80440.1 JAT86330.1 JAT90005.1 JTDY01006698 KOB65763.1 AGBW02012947 OWR44182.1 DS232245 EDS38326.1 GFDL01008250 JAV26795.1 GGFM01007354 MBW28105.1 GGFJ01007807 MBW56948.1 JTDY01000627 KOB76419.1 ADMH02000766 ETN65108.1 UFQT01000339 SSX23354.1 ATLV01027083 KE525423 KFB53854.1 AAAB01008848 EAA07027.4 APCN01003547 JTDY01006610 KOB65836.1 GGFK01008332 MBW41653.1 GAMD01002708 JAA98882.1 AXCN02001814 AXCM01001000 AJVK01014661 CH477190 EAT48696.1 GAPW01002874 JAC10724.1 GANO01002042 JAB57829.1 CP012524 ALC41872.1 CH933808 EDW10822.1 KK107459 QOIP01000004 EZA50393.1 RLU24234.1 GL436748 EFN71540.1 GAKP01007552 JAC51400.1 AJWK01005748 AJWK01005749 LBMM01002768 KMQ94397.1 GAMC01007708 JAB98847.1 KV925179 PIO35124.1 GL452364 EFN77399.1 GBXI01013035 JAD01257.1 AE013599 AY061358 GAKP01007553 JAC51399.1 GDHF01014400 JAI37914.1 KQ976500 KYM83187.1 KQ981673 KYN38362.1 KQ980581 KYN15269.1 ADTU01011295 CH480820 EDW53946.1 CM000362 CM002911 EDX08518.1 KMY96290.1 GEBQ01023476 JAT16501.1 GECZ01000508 JAS69261.1 GEBQ01023192 JAT16785.1 CH902619 EDV37491.1 PYGN01000160 PSN52497.1 PIO35125.1 JRES01000857 KNC27715.1 GEZM01042433 JAV79854.1 GECZ01029236 GECZ01020891 GECZ01003524 JAS40533.1 JAS48878.1 JAS66245.1 RJVU01048900 ROL43072.1 CCAG010006200 LJIG01009893 KRT82170.1 CM000158 EDW92502.1 KQ982373 KYQ57009.1 KQ971338 EFA01792.1 CH940648 EDW60630.1 GBBI01003815 JAC14897.1 GFDF01002777 JAV11307.1 NNAY01000467 OXU28168.1 GDKW01001904 JAI54691.1 IACK01139303 LAA88665.1 GFDF01002776 JAV11308.1 IACK01139301 LAA88662.1 GEBQ01029413 JAT10564.1 GEDC01027265 GEDC01026000 JAS10033.1 JAS11298.1 CH916367 EDW00620.1 GEBQ01005298 JAT34679.1 EZ423211 ADD19487.1 JXJN01022766
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000053268
UP000037510
+ More
UP000007151 UP000069272 UP000002320 UP000075881 UP000000673 UP000075885 UP000075880 UP000030765 UP000075882 UP000075903 UP000076407 UP000007062 UP000075840 UP000075902 UP000075884 UP000076408 UP000075886 UP000075883 UP000075920 UP000075901 UP000075900 UP000092462 UP000008820 UP000092553 UP000009192 UP000053097 UP000279307 UP000000311 UP000092461 UP000095300 UP000036403 UP000008237 UP000000803 UP000078540 UP000078541 UP000078492 UP000005205 UP000001292 UP000000304 UP000007801 UP000245037 UP000037069 UP000092445 UP000078200 UP000092444 UP000002282 UP000192221 UP000075809 UP000007266 UP000008792 UP000215335 UP000002358 UP000005203 UP000001070 UP000245300 UP000002280 UP000092460
UP000007151 UP000069272 UP000002320 UP000075881 UP000000673 UP000075885 UP000075880 UP000030765 UP000075882 UP000075903 UP000076407 UP000007062 UP000075840 UP000075902 UP000075884 UP000076408 UP000075886 UP000075883 UP000075920 UP000075901 UP000075900 UP000092462 UP000008820 UP000092553 UP000009192 UP000053097 UP000279307 UP000000311 UP000092461 UP000095300 UP000036403 UP000008237 UP000000803 UP000078540 UP000078541 UP000078492 UP000005205 UP000001292 UP000000304 UP000007801 UP000245037 UP000037069 UP000092445 UP000078200 UP000092444 UP000002282 UP000192221 UP000075809 UP000007266 UP000008792 UP000215335 UP000002358 UP000005203 UP000001070 UP000245300 UP000002280 UP000092460
Interpro
Gene 3D
ProteinModelPortal
H9IV38
A0A2A4JCN7
A0A2W1C271
A0A194RDF8
A0A2H1VTT7
A0A3S2PIF6
+ More
A0A194Q6L2 S4PX53 A0A1E1WSJ8 A0A0L7KR62 A0A212ERR6 A0A182FVZ2 B0X0T4 A0A182JQ60 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 A0A0L7LM38 W5JP62 A0A182PN54 A0A336LZD2 A0A182J590 A0A084WUG0 A0A182KUZ0 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A0L7KRZ7 A0A2M4ALI7 T1E8J9 A0A182UBB7 A0A182NPA1 A0A182YAB5 A0A182Q2A3 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182RLV4 A0A1B0DDY6 Q17PQ5 A0A023EQ93 U5EYF0 A0A0M4EH73 B4KM33 A0A026W2Z4 E2A4U6 A0A034WB86 A0A1B0CBQ1 A0A1I8NLH3 A0A0J7KV79 W8BJC5 A0A2G9S4V8 E2C413 A0A0A1WQY6 Q9W197 A0A034W9L4 A0A0K8VG61 A0A195BG84 A0A195FDT2 A0A195DRX9 A0A158NBS5 B4I2F1 B4QBK0 A0A1B6KYL7 A0A1B6H3L6 A0A1B6KZG4 B3MDM8 A0A2P8Z7N4 A0A2G9S4X1 A0A0L0C605 A0A1Y1M1Z2 A0A1B6FFA8 A0A3N0YA27 A0A1A9ZQS9 A0A1A9UER1 A0A1B0FE50 A0A0T6B439 B4PAW2 A0A1W4UM40 A0A151X9L3 D1ZZV3 B4LK41 A0A023F065 A0A1L8DXW8 A0A232FCJ5 K7J061 A0A088ATS9 A0A0P4VK67 A0A2D4IWN5 A0A1L8DXY4 A0A2D4IWS6 A0A1B6KHD2 A0A1B6C974 B4J4S4 A0A1B6MFL6 S4RFV0 F7DEV4 D3TP59 A0A1B0BYQ4
A0A194Q6L2 S4PX53 A0A1E1WSJ8 A0A0L7KR62 A0A212ERR6 A0A182FVZ2 B0X0T4 A0A182JQ60 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 A0A0L7LM38 W5JP62 A0A182PN54 A0A336LZD2 A0A182J590 A0A084WUG0 A0A182KUZ0 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A0L7KRZ7 A0A2M4ALI7 T1E8J9 A0A182UBB7 A0A182NPA1 A0A182YAB5 A0A182Q2A3 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182RLV4 A0A1B0DDY6 Q17PQ5 A0A023EQ93 U5EYF0 A0A0M4EH73 B4KM33 A0A026W2Z4 E2A4U6 A0A034WB86 A0A1B0CBQ1 A0A1I8NLH3 A0A0J7KV79 W8BJC5 A0A2G9S4V8 E2C413 A0A0A1WQY6 Q9W197 A0A034W9L4 A0A0K8VG61 A0A195BG84 A0A195FDT2 A0A195DRX9 A0A158NBS5 B4I2F1 B4QBK0 A0A1B6KYL7 A0A1B6H3L6 A0A1B6KZG4 B3MDM8 A0A2P8Z7N4 A0A2G9S4X1 A0A0L0C605 A0A1Y1M1Z2 A0A1B6FFA8 A0A3N0YA27 A0A1A9ZQS9 A0A1A9UER1 A0A1B0FE50 A0A0T6B439 B4PAW2 A0A1W4UM40 A0A151X9L3 D1ZZV3 B4LK41 A0A023F065 A0A1L8DXW8 A0A232FCJ5 K7J061 A0A088ATS9 A0A0P4VK67 A0A2D4IWN5 A0A1L8DXY4 A0A2D4IWS6 A0A1B6KHD2 A0A1B6C974 B4J4S4 A0A1B6MFL6 S4RFV0 F7DEV4 D3TP59 A0A1B0BYQ4
PDB
4NWI
E-value=1.64802e-08,
Score=134
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
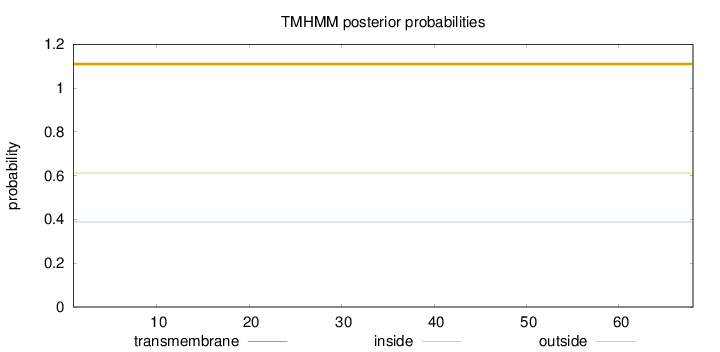
Length:
68
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.38775
outside
1 - 68
Population Genetic Test Statistics
Pi
11.601708
Theta
9.146509
Tajima's D
0.783046
CLR
0.221912
CSRT
0.613319334033298
Interpretation
Uncertain
