Gene
KWMTBOMO08060
Pre Gene Modal
BGIBMGA001118
Annotation
PREDICTED:_7-methylguanosine_phosphate-specific_5'-nucleotidase_[Papilio_machaon]
Full name
5'-nucleotidase
Location in the cell
Cytoplasmic Reliability : 1.717 Extracellular Reliability : 1.073
Sequence
CDS
ATGATAGACAGTTTTAGGAAGGGCGTTAAAGAATTGATAATTTGGAGTCAGACCCATGAGGTTCCGGTGCTGGTGTTCTCGGCGGGATTAGGAGAATCCGTTGTGGCAGCTTTGAAGGCGGCGAATTTTTTGTTACCGCACGTCAAGGTCGTATCAAACTTCTTAGCTCTCGACGAGAACGATAAAATAGTCGGTATAAAAGGCGAGGTCATACACACGTACAACAAAAACGAAACTGCCATTAAAAACACCGATTATTATAATTTAGTACAGCAAAGGAGTAATGTCATCTTAATGGGGGATAATATCGGTGACGCCGGAATGGCTGAGGGAATGGACCATTGTGATGTCGTCGTCAAAGTAGGGTTCCTCGGTCATAGCGCCGAAGGGAACCTCCCTAACTACAAGAAGAAATTCGATATAGTCCTAGTCAACGAACCATCTATGGACATTGTCAATGCCATGGTGAAACTGGTGCTGTGA
Protein
MIDSFRKGVKELIIWSQTHEVPVLVFSAGLGESVVAALKAANFLLPHVKVVSNFLALDENDKIVGIKGEVIHTYNKNETAIKNTDYYNLVQQRSNVILMGDNIGDAGMAEGMDHCDVVVKVGFLGHSAEGNLPNYKKKFDIVLVNEPSMDIVNAMVKLVL
Summary
Catalytic Activity
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
Similarity
Belongs to the pyrimidine 5'-nucleotidase family.
Uniprot
H9IV38
A0A1E1WSJ8
A0A2A4JCN7
A0A2W1C271
A0A194RDF8
A0A194Q6L2
+ More
A0A2H1VTT7 S4PX53 A0A212ERR6 A0A3S2PIF6 A0A0L7KR62 D1ZZV3 A0A0L7KRZ7 A0A0L7LM38 Q17PQ4 Q17PQ5 A0A023EQ93 A0A182GPQ4 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 T1E8J9 B0X0T4 A0A1W4XW98 N6TEJ5 W5JP62 U5EYF0 A0A182RLV4 A0A2M4ALI7 A0A1L8DXY4 A0A1Y1M1Z2 A0A084WUG0 A0A1L8DXW8 A0A182FVZ2 A0A182UBB7 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A182J590 A0A182KUZ0 A0A2P8Z7M7 A0A1B0CBQ1 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182YAB5 A0A182JQ60 A0A182PN54 A0A182NPA1 A0A2C9JMN7 A0A2C9JMQ4 A0A182Q2A3 A0A2J7QMJ2 A0A067RFQ8 A0A0K8V8A8 A0A1Y1M6C8 W4Y0K4 A0A0K8WAW8 A0A146LXQ4 A0A0K8SWY5 A0A3B0J2N6 A0A336N2K4 A0A2L2Y8B5 A0A0S7GZ64 A0A0B6Z9L2 G3NA97 Q28WT6 B4H6A2 A0A1B6C974 A0A3B4XNG7 A0A3B4TAU3 A0A3B4B262 A0A336LZD2 A0A0M4EH73 A0A0K8VG61 A0A034W9L4 A0A3P8NMB8 A0A3P9BT24 A0A3B3CE63 A0A3B4B3A1 A0A3B5B6C9 A0A2P2I1C9 A0A3B4XLN1 A0A1W4UM40 A0A3Q3M5Y6 B3NQG7 A0A3Q3M0E4 B4J4S4 V9KNI6 A0A3Q2Y0D1 A0A2U9CGM2 C3YZ54 A0A3P8VIW4 A0A2D0T6C7 I3JHX0 V9KK26 B4MRD4 W8BJC5 B4LK41
A0A2H1VTT7 S4PX53 A0A212ERR6 A0A3S2PIF6 A0A0L7KR62 D1ZZV3 A0A0L7KRZ7 A0A0L7LM38 Q17PQ4 Q17PQ5 A0A023EQ93 A0A182GPQ4 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 T1E8J9 B0X0T4 A0A1W4XW98 N6TEJ5 W5JP62 U5EYF0 A0A182RLV4 A0A2M4ALI7 A0A1L8DXY4 A0A1Y1M1Z2 A0A084WUG0 A0A1L8DXW8 A0A182FVZ2 A0A182UBB7 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A182J590 A0A182KUZ0 A0A2P8Z7M7 A0A1B0CBQ1 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182YAB5 A0A182JQ60 A0A182PN54 A0A182NPA1 A0A2C9JMN7 A0A2C9JMQ4 A0A182Q2A3 A0A2J7QMJ2 A0A067RFQ8 A0A0K8V8A8 A0A1Y1M6C8 W4Y0K4 A0A0K8WAW8 A0A146LXQ4 A0A0K8SWY5 A0A3B0J2N6 A0A336N2K4 A0A2L2Y8B5 A0A0S7GZ64 A0A0B6Z9L2 G3NA97 Q28WT6 B4H6A2 A0A1B6C974 A0A3B4XNG7 A0A3B4TAU3 A0A3B4B262 A0A336LZD2 A0A0M4EH73 A0A0K8VG61 A0A034W9L4 A0A3P8NMB8 A0A3P9BT24 A0A3B3CE63 A0A3B4B3A1 A0A3B5B6C9 A0A2P2I1C9 A0A3B4XLN1 A0A1W4UM40 A0A3Q3M5Y6 B3NQG7 A0A3Q3M0E4 B4J4S4 V9KNI6 A0A3Q2Y0D1 A0A2U9CGM2 C3YZ54 A0A3P8VIW4 A0A2D0T6C7 I3JHX0 V9KK26 B4MRD4 W8BJC5 B4LK41
EC Number
3.1.3.5
Pubmed
19121390
28756777
26354079
23622113
22118469
26227816
+ More
18362917 19820115 17510324 24945155 26483478 23537049 20920257 23761445 28004739 24438588 12364791 14747013 17210077 20966253 29403074 25244985 15562597 24845553 26823975 26561354 15632085 17994087 25463417 25348373 25186727 29451363 24402279 18563158 24487278 24495485
18362917 19820115 17510324 24945155 26483478 23537049 20920257 23761445 28004739 24438588 12364791 14747013 17210077 20966253 29403074 25244985 15562597 24845553 26823975 26561354 15632085 17994087 25463417 25348373 25186727 29451363 24402279 18563158 24487278 24495485
EMBL
BABH01000388
GDQN01010614
GDQN01004724
GDQN01001049
JAT80440.1
JAT86330.1
+ More
JAT90005.1 NWSH01001864 PCG69867.1 KZ149891 PZC79076.1 KQ460397 KPJ15315.1 KQ459439 KPJ01014.1 ODYU01004396 SOQ44230.1 GAIX01005638 JAA86922.1 AGBW02012947 OWR44182.1 RSAL01000023 RVE52277.1 JTDY01006698 KOB65763.1 KQ971338 EFA01792.1 JTDY01006610 KOB65836.1 JTDY01000627 KOB76419.1 CH477190 EAT48697.1 EAT48696.1 GAPW01002874 JAC10724.1 JXUM01079005 KQ563101 KXJ74460.1 GFDL01008250 JAV26795.1 GGFM01007354 MBW28105.1 GGFJ01007807 MBW56948.1 GAMD01002708 JAA98882.1 DS232245 EDS38326.1 APGK01032103 APGK01032104 KB740835 KB632168 ENN78794.1 ERL89388.1 ADMH02000766 ETN65108.1 GANO01002042 JAB57829.1 GGFK01008332 MBW41653.1 GFDF01002776 JAV11308.1 GEZM01042433 JAV79854.1 ATLV01027083 KE525423 KFB53854.1 GFDF01002777 JAV11307.1 AAAB01008848 EAA07027.4 APCN01003547 PYGN01000160 PSN52496.1 AJWK01005748 AJWK01005749 AXCM01001000 AXCN02001814 NEVH01013205 PNF29815.1 KK852498 KDR22681.1 GDHF01017145 JAI35169.1 JAV79855.1 AAGJ04150365 AAGJ04150366 AAGJ04150367 AAGJ04150368 AAGJ04150369 AAGJ04150370 GDHF01004314 JAI48000.1 GDHC01006276 JAQ12353.1 GBRD01008155 JAG57666.1 OUUW01000001 SPP73402.1 UFQT01004973 SSX35839.1 IAAA01017168 IAAA01017169 IAAA01017170 LAA03570.1 GBYX01448962 GBYX01448961 JAO32521.1 HACG01018353 CEK65218.1 CM000071 EAL26580.1 CH479213 EDW33326.1 GEDC01027265 GEDC01026000 JAS10033.1 JAS11298.1 UFQT01000339 SSX23354.1 CP012524 ALC41872.1 GDHF01014400 JAI37914.1 GAKP01007553 JAC51399.1 IACF01002182 LAB67845.1 CH954179 EDV56970.1 CH916367 EDW00620.1 JW867077 AFO99594.1 CP026258 AWP15751.1 GG666565 EEN54562.1 AERX01024054 AERX01024055 AERX01024056 AERX01024057 JW865857 AFO98374.1 CH963850 EDW74673.1 GAMC01007708 JAB98847.1 CH940648 EDW60630.1
JAT90005.1 NWSH01001864 PCG69867.1 KZ149891 PZC79076.1 KQ460397 KPJ15315.1 KQ459439 KPJ01014.1 ODYU01004396 SOQ44230.1 GAIX01005638 JAA86922.1 AGBW02012947 OWR44182.1 RSAL01000023 RVE52277.1 JTDY01006698 KOB65763.1 KQ971338 EFA01792.1 JTDY01006610 KOB65836.1 JTDY01000627 KOB76419.1 CH477190 EAT48697.1 EAT48696.1 GAPW01002874 JAC10724.1 JXUM01079005 KQ563101 KXJ74460.1 GFDL01008250 JAV26795.1 GGFM01007354 MBW28105.1 GGFJ01007807 MBW56948.1 GAMD01002708 JAA98882.1 DS232245 EDS38326.1 APGK01032103 APGK01032104 KB740835 KB632168 ENN78794.1 ERL89388.1 ADMH02000766 ETN65108.1 GANO01002042 JAB57829.1 GGFK01008332 MBW41653.1 GFDF01002776 JAV11308.1 GEZM01042433 JAV79854.1 ATLV01027083 KE525423 KFB53854.1 GFDF01002777 JAV11307.1 AAAB01008848 EAA07027.4 APCN01003547 PYGN01000160 PSN52496.1 AJWK01005748 AJWK01005749 AXCM01001000 AXCN02001814 NEVH01013205 PNF29815.1 KK852498 KDR22681.1 GDHF01017145 JAI35169.1 JAV79855.1 AAGJ04150365 AAGJ04150366 AAGJ04150367 AAGJ04150368 AAGJ04150369 AAGJ04150370 GDHF01004314 JAI48000.1 GDHC01006276 JAQ12353.1 GBRD01008155 JAG57666.1 OUUW01000001 SPP73402.1 UFQT01004973 SSX35839.1 IAAA01017168 IAAA01017169 IAAA01017170 LAA03570.1 GBYX01448962 GBYX01448961 JAO32521.1 HACG01018353 CEK65218.1 CM000071 EAL26580.1 CH479213 EDW33326.1 GEDC01027265 GEDC01026000 JAS10033.1 JAS11298.1 UFQT01000339 SSX23354.1 CP012524 ALC41872.1 GDHF01014400 JAI37914.1 GAKP01007553 JAC51399.1 IACF01002182 LAB67845.1 CH954179 EDV56970.1 CH916367 EDW00620.1 JW867077 AFO99594.1 CP026258 AWP15751.1 GG666565 EEN54562.1 AERX01024054 AERX01024055 AERX01024056 AERX01024057 JW865857 AFO98374.1 CH963850 EDW74673.1 GAMC01007708 JAB98847.1 CH940648 EDW60630.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000037510 UP000007266 UP000008820 UP000069940 UP000249989 UP000002320 UP000192223 UP000019118 UP000030742 UP000000673 UP000075900 UP000030765 UP000069272 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000075880 UP000075882 UP000245037 UP000092461 UP000075883 UP000075920 UP000075901 UP000076408 UP000075881 UP000075885 UP000075884 UP000076420 UP000075886 UP000235965 UP000027135 UP000007110 UP000268350 UP000007635 UP000001819 UP000008744 UP000261360 UP000261420 UP000261520 UP000092553 UP000265100 UP000265160 UP000261560 UP000261400 UP000192221 UP000261660 UP000008711 UP000261640 UP000001070 UP000264820 UP000246464 UP000001554 UP000265120 UP000221080 UP000005207 UP000007798 UP000008792
UP000037510 UP000007266 UP000008820 UP000069940 UP000249989 UP000002320 UP000192223 UP000019118 UP000030742 UP000000673 UP000075900 UP000030765 UP000069272 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000075880 UP000075882 UP000245037 UP000092461 UP000075883 UP000075920 UP000075901 UP000076408 UP000075881 UP000075885 UP000075884 UP000076420 UP000075886 UP000235965 UP000027135 UP000007110 UP000268350 UP000007635 UP000001819 UP000008744 UP000261360 UP000261420 UP000261520 UP000092553 UP000265100 UP000265160 UP000261560 UP000261400 UP000192221 UP000261660 UP000008711 UP000261640 UP000001070 UP000264820 UP000246464 UP000001554 UP000265120 UP000221080 UP000005207 UP000007798 UP000008792
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IV38
A0A1E1WSJ8
A0A2A4JCN7
A0A2W1C271
A0A194RDF8
A0A194Q6L2
+ More
A0A2H1VTT7 S4PX53 A0A212ERR6 A0A3S2PIF6 A0A0L7KR62 D1ZZV3 A0A0L7KRZ7 A0A0L7LM38 Q17PQ4 Q17PQ5 A0A023EQ93 A0A182GPQ4 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 T1E8J9 B0X0T4 A0A1W4XW98 N6TEJ5 W5JP62 U5EYF0 A0A182RLV4 A0A2M4ALI7 A0A1L8DXY4 A0A1Y1M1Z2 A0A084WUG0 A0A1L8DXW8 A0A182FVZ2 A0A182UBB7 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A182J590 A0A182KUZ0 A0A2P8Z7M7 A0A1B0CBQ1 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182YAB5 A0A182JQ60 A0A182PN54 A0A182NPA1 A0A2C9JMN7 A0A2C9JMQ4 A0A182Q2A3 A0A2J7QMJ2 A0A067RFQ8 A0A0K8V8A8 A0A1Y1M6C8 W4Y0K4 A0A0K8WAW8 A0A146LXQ4 A0A0K8SWY5 A0A3B0J2N6 A0A336N2K4 A0A2L2Y8B5 A0A0S7GZ64 A0A0B6Z9L2 G3NA97 Q28WT6 B4H6A2 A0A1B6C974 A0A3B4XNG7 A0A3B4TAU3 A0A3B4B262 A0A336LZD2 A0A0M4EH73 A0A0K8VG61 A0A034W9L4 A0A3P8NMB8 A0A3P9BT24 A0A3B3CE63 A0A3B4B3A1 A0A3B5B6C9 A0A2P2I1C9 A0A3B4XLN1 A0A1W4UM40 A0A3Q3M5Y6 B3NQG7 A0A3Q3M0E4 B4J4S4 V9KNI6 A0A3Q2Y0D1 A0A2U9CGM2 C3YZ54 A0A3P8VIW4 A0A2D0T6C7 I3JHX0 V9KK26 B4MRD4 W8BJC5 B4LK41
A0A2H1VTT7 S4PX53 A0A212ERR6 A0A3S2PIF6 A0A0L7KR62 D1ZZV3 A0A0L7KRZ7 A0A0L7LM38 Q17PQ4 Q17PQ5 A0A023EQ93 A0A182GPQ4 A0A1Q3FGY6 A0A2M3ZI08 A0A2M4BW44 T1E8J9 B0X0T4 A0A1W4XW98 N6TEJ5 W5JP62 U5EYF0 A0A182RLV4 A0A2M4ALI7 A0A1L8DXY4 A0A1Y1M1Z2 A0A084WUG0 A0A1L8DXW8 A0A182FVZ2 A0A182UBB7 A0A182UNK3 A0A182WY01 Q7QE29 A0A182I621 A0A182J590 A0A182KUZ0 A0A2P8Z7M7 A0A1B0CBQ1 A0A182LXU3 A0A182WE85 A0A182S7E1 A0A182YAB5 A0A182JQ60 A0A182PN54 A0A182NPA1 A0A2C9JMN7 A0A2C9JMQ4 A0A182Q2A3 A0A2J7QMJ2 A0A067RFQ8 A0A0K8V8A8 A0A1Y1M6C8 W4Y0K4 A0A0K8WAW8 A0A146LXQ4 A0A0K8SWY5 A0A3B0J2N6 A0A336N2K4 A0A2L2Y8B5 A0A0S7GZ64 A0A0B6Z9L2 G3NA97 Q28WT6 B4H6A2 A0A1B6C974 A0A3B4XNG7 A0A3B4TAU3 A0A3B4B262 A0A336LZD2 A0A0M4EH73 A0A0K8VG61 A0A034W9L4 A0A3P8NMB8 A0A3P9BT24 A0A3B3CE63 A0A3B4B3A1 A0A3B5B6C9 A0A2P2I1C9 A0A3B4XLN1 A0A1W4UM40 A0A3Q3M5Y6 B3NQG7 A0A3Q3M0E4 B4J4S4 V9KNI6 A0A3Q2Y0D1 A0A2U9CGM2 C3YZ54 A0A3P8VIW4 A0A2D0T6C7 I3JHX0 V9KK26 B4MRD4 W8BJC5 B4LK41
PDB
4NWI
E-value=1.78723e-31,
Score=334
Ontologies
GO
PANTHER
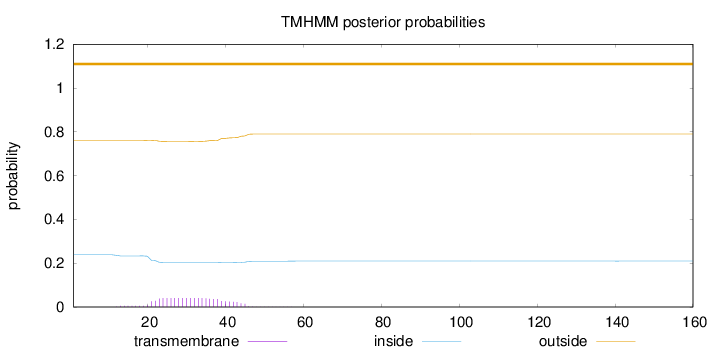
Topology
Subcellular location
Cytoplasm
Length:
160
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.900569999999998
Exp number, first 60 AAs:
0.89924
Total prob of N-in:
0.23810
outside
1 - 160
Population Genetic Test Statistics
Pi
14.848174
Theta
13.977292
Tajima's D
-0.94776
CLR
0.873013
CSRT
0.145292735363232
Interpretation
Uncertain
