Gene
KWMTBOMO08059
Pre Gene Modal
BGIBMGA001042
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_probable_phospholipid-transporting_ATPase_VD_[Bombyx_mori]
Full name
Phospholipid-transporting ATPase
+ More
P-type phospholipid transporter
P-type phospholipid transporter
Location in the cell
PlasmaMembrane Reliability : 2.653
Sequence
CDS
ATGCTTGCATGTTTGTGTTACAGCGACATTAGTAGATCAGCCGAAAGATATGTTAAGGTGAAATGGAGGGACGTTCGAGTCGGCGACCTCGTTCACTTGTCTAACAACGAAGCGGTGCCAGCTGATATGGTGCTGCTGCACTCCTCCAATCCAATGGGGATCTGCTACCTGGACACGTGTAATCTGGACGGAGAGACAAACTTGAAACAGAGAATCGTCGCACCGGGCTTCAGAGAAAAGCGACTTGATTTTAATCCAATGAAATTCAGAAGTTCAATAGAAGTCGAAAGGCCTTCAACGAAAATCTACAGGGAATGCACGATCAAGAACACGGATTACGTCGAGGGTATCGTTGTGTACGCGGGACACGAGACCAAAGCCATGCTGAACAACGGGGGACCGCGATACAAATGCTCCGGTCTGGAGAAGAAAATGAACACAGACGTGATTTGGTGTGTTTTAGTGCTGCTATTCCTCTGCTGCGTTGGCGCCGCCTGTTACAAAGTCTGGTTGGACCAATACTTTCCCAATTTACTAAGATACACGCTTTTCATTCCGCAAGCTGACAAACCACCGGCGTACGAAGGCCTACAGATATTCTGGACCTACATTATAATATTACAAGTCATGATACCAATATCTCTGTACGTGACTATTGAGATGACGAAACTATTGCAAGTCTATCATATACATCAAGATATAGAAATGTACGATCCGGAGACGAACATCAGAACCGAATGCCGAGCGCTCAACATAACGGAGGAACTGGGACAGATAAGTTACTTGTTCAGCGATAAGACCGGAACGTTGACCGAGAACAAGATGGTGTTTAGGAGATGCACCGTTAACGGTGTCGATTACGATCACCCGCCTGGACCCCCCGCGGAACCCTCGACCGAACTCCCGCCCATTGTAACTCCTCTAACCCAAGTATCGCCAAACCGAAGACTGCTCCAACATCTAAACGACACAAACGATGCTCAACACACGCAAAAGGTTCGCGAGTTCCTCTTGATCTTGGCCGTCTGCAATACGGTGGTCGTGAGTCAGCCTCACGTGGACACGATGCAGCTCAGCATGTCGAATTCGGGCGAAGAACTGACGAACAACAGGAAACCATCGAGATCGAATGGTACATTACGATCAAACGACAAGTACGCTCGATTAACGGAATCGCGATCGACGACGCCCTCGCCACCGCCCTCGACCGTTTCCCCCCTGAAGATACGTCTGCCTAAACTGACGTTTGGTAGCAGAGACGACAACACCAGTGAACCGAGTACATCTACTGAACAACAGATCGCTCGTTTTGAAGCGGAAAGCCCGGACGAGCTAGCACTCGCTGAGGCGGCTCTCGCGTACGGCTACGAGCTGAGAAGCAGGTCGCCTGACGAGGTCGAGGTCGGGATCAAGGGGGAGGTGAGCCGCCTGAAGGTGCTTCGGGTGCAGCAGTTCGACTCCAACCGGAAATGCATGTCCGTCGCGATGCGGACGCCTACCGGTCAAGTAGTTTTGTACGTAAAAGGCGCTGATAGCACAGTGCTGGGCGCCTTGGCACCGATGCGGGCCGGCTCGGCGGAGGCGGCGGCGTGCGAGCGCACCCGCGCCCTCATCAGCGAATACTCGCGCGCCGGCCTCCGCACGCTGGTCATGGCCAAGCGCACCATGCAGCCCGCGCTGTGGGAGGAGTGGCTCGCCAGCCATACCAGAGCCTCGGAGATCGGAGAAGGCCGCGATAAGAGGATACGCGACTCGCTGACTCGGCTCGAGAGTGCCCTGACGCTGGTGGGGGCCACGGGGGTCGAGGACCGGCTGCAGGAGGACGTGCCGCGCACCGTCCGGGCGCTGCTCGACGCCGGGATCATCGTGTGGGTGCTGACCGGGGACAAGCCGGAGACCGCCATCAACATAGCGTACTCCGCCTCTCTGTTCTCACAGAGCGACCGGCTGTTGCACCTCATGTCCAGGGATAAGGAGCACGCGGAGTCCACGATCAAGAGTTACCTGGAGGGGGGTGTGGGGGGTGCGGGGCGCGCGCTGGTGGTGGACGGGCGCACGCTCACGTACATCCTGGACCGGCGCTCGGGGCTGGTGGCGCCGTTCCTGTGTCTGGCGCGGCGCTGCTCCGCCGTGCTGTGCTGCCGCGCCACGCCGCTGCAGAAGGCCTGCATCGTCAAGGCGGTGAAGGAAGAGCTCGGCGTGACGACCCTGGCCATAGGCGACGGCGCCAACGACGTCTCCATGATACAAACGGCTGACGTCGGCGTCGGTCTGTCTGGCCAGGAAGGTATGCAGGCGGTGATGGCTTCGGACTTCGCGCTGTCGAGGTTCAAGTTCATAGAGCGGCTGCTGCTCGTACACGGACACTGGTGCTATGACCGGCTCGCCAGGATGATACTGTATTTCTTCCTCAAAAATGCCACGTTCGTGTTCATGGTGTTCTGGTACCAGCTGTACTGCGGCTTCTCAGCTACGGTGATGATCGACCAGCTACACCTCATGGCCTACAACCTCATGTTCACCGCGCTACCACCCATCGTCATCGGTAAGCTCTCATCTACTCCAAACTTATAA
Protein
MLACLCYSDISRSAERYVKVKWRDVRVGDLVHLSNNEAVPADMVLLHSSNPMGICYLDTCNLDGETNLKQRIVAPGFREKRLDFNPMKFRSSIEVERPSTKIYRECTIKNTDYVEGIVVYAGHETKAMLNNGGPRYKCSGLEKKMNTDVIWCVLVLLFLCCVGAACYKVWLDQYFPNLLRYTLFIPQADKPPAYEGLQIFWTYIIILQVMIPISLYVTIEMTKLLQVYHIHQDIEMYDPETNIRTECRALNITEELGQISYLFSDKTGTLTENKMVFRRCTVNGVDYDHPPGPPAEPSTELPPIVTPLTQVSPNRRLLQHLNDTNDAQHTQKVREFLLILAVCNTVVVSQPHVDTMQLSMSNSGEELTNNRKPSRSNGTLRSNDKYARLTESRSTTPSPPPSTVSPLKIRLPKLTFGSRDDNTSEPSTSTEQQIARFEAESPDELALAEAALAYGYELRSRSPDEVEVGIKGEVSRLKVLRVQQFDSNRKCMSVAMRTPTGQVVLYVKGADSTVLGALAPMRAGSAEAAACERTRALISEYSRAGLRTLVMAKRTMQPALWEEWLASHTRASEIGEGRDKRIRDSLTRLESALTLVGATGVEDRLQEDVPRTVRALLDAGIIVWVLTGDKPETAINIAYSASLFSQSDRLLHLMSRDKEHAESTIKSYLEGGVGGAGRALVVDGRTLTYILDRRSGLVAPFLCLARRCSAVLCCRATPLQKACIVKAVKEELGVTTLAIGDGANDVSMIQTADVGVGLSGQEGMQAVMASDFALSRFKFIERLLLVHGHWCYDRLARMILYFFLKNATFVFMVFWYQLYCGFSATVMIDQLHLMAYNLMFTALPPIVIGKLSSTPNL
Summary
Catalytic Activity
ATP + H(2)O + phospholipid(Side 1) = ADP + phosphate + phospholipid(Side 2).
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IV subfamily.
Uniprot
H9IUW2
A0A212ERV6
A0A2A4K0N8
A0A194RCX0
A0A3S2NJ37
A0A2H1VTS1
+ More
A0A2W1C037 A0A194Q650 U4U0G1 A0A336KFC8 A0A336KDK6 A0A3Q3LHG2 A0A3B4XIT5 A0A3Q2XHL2 A0A2B4SW24 A0A3B3WXE1 A0A3P9NFD8 A0A3P9NFC5 A0A3Q1AZZ1 A0A3P8P071 A0A315VKE5 A0A3Q0RBZ5 A0A3B4EPK4 A0A3B5LL55 A0A3Q2D9L1 M4A4G4 A0A3B5AEX2 A0A2I4B991 A0A2I4B985 A0A3Q3F2P1 A0A0J7L9T1 A0A3P8TT13
A0A2W1C037 A0A194Q650 U4U0G1 A0A336KFC8 A0A336KDK6 A0A3Q3LHG2 A0A3B4XIT5 A0A3Q2XHL2 A0A2B4SW24 A0A3B3WXE1 A0A3P9NFD8 A0A3P9NFC5 A0A3Q1AZZ1 A0A3P8P071 A0A315VKE5 A0A3Q0RBZ5 A0A3B4EPK4 A0A3B5LL55 A0A3Q2D9L1 M4A4G4 A0A3B5AEX2 A0A2I4B991 A0A2I4B985 A0A3Q3F2P1 A0A0J7L9T1 A0A3P8TT13
EC Number
7.6.2.1
EMBL
BABH01000390
BABH01000391
AGBW02012947
OWR44181.1
NWSH01000278
PCG77807.1
+ More
KQ460397 KPJ15314.1 RSAL01000023 RVE52278.1 ODYU01004396 SOQ44229.1 KZ149891 PZC79075.1 KQ459439 KPJ01012.1 KB631829 ERL86572.1 UFQS01000300 UFQT01000300 SSX02612.1 SSX22986.1 SSX02611.1 SSX22985.1 LSMT01000016 PFX33080.1 NHOQ01001578 PWA23561.1 LBMM01000107 KMR04921.1
KQ460397 KPJ15314.1 RSAL01000023 RVE52278.1 ODYU01004396 SOQ44229.1 KZ149891 PZC79075.1 KQ459439 KPJ01012.1 KB631829 ERL86572.1 UFQS01000300 UFQT01000300 SSX02612.1 SSX22986.1 SSX02611.1 SSX22985.1 LSMT01000016 PFX33080.1 NHOQ01001578 PWA23561.1 LBMM01000107 KMR04921.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000283053
UP000053268
+ More
UP000030742 UP000261640 UP000261360 UP000264820 UP000225706 UP000261480 UP000242638 UP000257160 UP000265100 UP000261340 UP000261460 UP000261380 UP000265020 UP000002852 UP000261400 UP000192220 UP000264800 UP000036403 UP000265080
UP000030742 UP000261640 UP000261360 UP000264820 UP000225706 UP000261480 UP000242638 UP000257160 UP000265100 UP000261340 UP000261460 UP000261380 UP000265020 UP000002852 UP000261400 UP000192220 UP000264800 UP000036403 UP000265080
PRIDE
Interpro
IPR018303
ATPase_P-typ_P_site
+ More
IPR032630 P_typ_ATPase_c
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR006539 P-type_ATPase_IV
IPR023299 ATPase_P-typ_cyto_dom_N
IPR001757 P_typ_ATPase
IPR023298 ATPase_P-typ_TM_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR032631 P-type_ATPase_N
IPR030359 ATP10B
IPR021134 Bestrophin/UPF0187
IPR032630 P_typ_ATPase_c
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR006539 P-type_ATPase_IV
IPR023299 ATPase_P-typ_cyto_dom_N
IPR001757 P_typ_ATPase
IPR023298 ATPase_P-typ_TM_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR032631 P-type_ATPase_N
IPR030359 ATP10B
IPR021134 Bestrophin/UPF0187
Gene 3D
ProteinModelPortal
H9IUW2
A0A212ERV6
A0A2A4K0N8
A0A194RCX0
A0A3S2NJ37
A0A2H1VTS1
+ More
A0A2W1C037 A0A194Q650 U4U0G1 A0A336KFC8 A0A336KDK6 A0A3Q3LHG2 A0A3B4XIT5 A0A3Q2XHL2 A0A2B4SW24 A0A3B3WXE1 A0A3P9NFD8 A0A3P9NFC5 A0A3Q1AZZ1 A0A3P8P071 A0A315VKE5 A0A3Q0RBZ5 A0A3B4EPK4 A0A3B5LL55 A0A3Q2D9L1 M4A4G4 A0A3B5AEX2 A0A2I4B991 A0A2I4B985 A0A3Q3F2P1 A0A0J7L9T1 A0A3P8TT13
A0A2W1C037 A0A194Q650 U4U0G1 A0A336KFC8 A0A336KDK6 A0A3Q3LHG2 A0A3B4XIT5 A0A3Q2XHL2 A0A2B4SW24 A0A3B3WXE1 A0A3P9NFD8 A0A3P9NFC5 A0A3Q1AZZ1 A0A3P8P071 A0A315VKE5 A0A3Q0RBZ5 A0A3B4EPK4 A0A3B5LL55 A0A3Q2D9L1 M4A4G4 A0A3B5AEX2 A0A2I4B991 A0A2I4B985 A0A3Q3F2P1 A0A0J7L9T1 A0A3P8TT13
PDB
6A69
E-value=7.53593e-12,
Score=173
Ontologies
GO
PANTHER
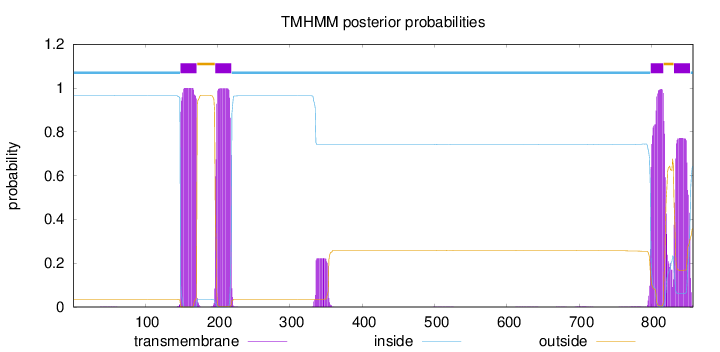
Topology
Subcellular location
Membrane
Length:
857
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
85.8393800000003
Exp number, first 60 AAs:
0.00596
Total prob of N-in:
0.96548
inside
1 - 148
TMhelix
149 - 171
outside
172 - 196
TMhelix
197 - 219
inside
220 - 798
TMhelix
799 - 816
outside
817 - 830
TMhelix
831 - 853
inside
854 - 857
Population Genetic Test Statistics
Pi
17.688628
Theta
15.668493
Tajima's D
0.260196
CLR
0.660245
CSRT
0.450077496125194
Interpretation
Uncertain
