Pre Gene Modal
BGIBMGA001038
Annotation
PREDICTED:_tudor_domain-containing_protein_3-like_[Bombyx_mori]
Full name
Tudor domain-containing protein 3
Location in the cell
Nuclear Reliability : 4.282
Sequence
CDS
ATGTCATTACGGGAAACTCTTAAGGAACTGGGCTGGCATTTGACCGAAGAAGGCATCGATACGATAACCGAAAATGGTCAAATTCAAGATGTAAACATTCTTAGCAAAAAAGCTTTAGACTACGATTTGAGAGACATCAGCGAAGGATCGTTACCTGAAGACTTTGTCAAGGATCCGTCTAAATTAGAAAGCCCCGTTGTTGTTCAAATTCAGAAGATTAGGAATGTGTCAGCTCCTAAAGCTAATGAAGAATCAACATCTGCCCCACGGATGTTGAAGCTAACCCTGCATGACGGGAAAGTAACGTGTACAGGTCTAGAGATAAGTCATATACCCAGTCTAAGTATAAACACCCCACCTGGAACTAAGTTGTTGCTGAAGAATGAAGAGCTAGAAGTTTGTCACGGAGTGGTATGGCTCACCCCATCAGTGATATCTGTATTGGGCGGAACAGTATCTCATATGATTGAAAAATGGGAACTGAACAGAAGTCTTGCCAAACACACAAGAGGAGGCATAGGAGCTGAAGGAGGTCCTCCACCATGGATTCCATTTGGACAGAGATTAGAAGCTCTTTCTGTGGACAAACAGTTCAAGAGTCTACAAGAAGCTGCCAAACAGGACAATCCAGAGTTCGAGGCTCAAAGAAAAGGTGCTATTGCTGAGGCACAGAGGATGTCTGGTGTGAAAAAGGTTTTTGGTGGTGGAACTAAACCTTTACTGGACGCAAATGTTCAAAAGATTGTAGATGCAGGGTTTACAGAGGAACAAGCAGAAGCAACATTAAAATATACCAAGAACAATGTTGATAGAGCACTAAAGATATTGCAGAAAAGAGACAACTCTGAAAATAGGAATAAAGAGAAACCACAGAAGGAACAAGATCCAGTGAAGCGTAAAGGTCGCCACAAAGAGTCTGGTGATGAGGATGGTGTGCCTGTGAAACCGTCAGGAAAAGTGTCATTGTTTGACTTCTTAGAAGATAAATTACCAAATGTACCAGAAAAAGATAAGAATACGCGACAAAATAACTCTAGTATGGAAGATAGATATGACAGAAGCCAAAATAGAGAAAGAAATAGTAATAGGAACCGACATGGCTCAAGACATGAAGGGGCCAACCGAAATCGGTCAGACCGACGCCATTATTCCAACGACAACTTGTATTCATCTTATAGAGAAGATAGAAAATATCAGTCGCAAAATGAGAAACCACCAAGATTTCAAAAGATACAAGAGGAGATGAACAAACATCATCAAAACATGAACATGCCATATAACAACACTTATCAACAAAATCAATACAAGGATAGACATATGAGAAACGATCGGAACCCAAATGATAGAGGTCCGCAACAAAATTATAATCACAATTCTAGTACAATGGATGGCTTGGTTGATGCTACATCCAAATTGAATCTATTATCAAACCAATCGAGAATAGATGAACAACAAGTGCATAACTACCAACAGCATACAGATCAAAGCTACCAATCTAAGCAAACCTATCCATCAAATCATAGTCAAGATATACCACCATTCAGAAGGAGTAATGATTTACATCAAGACGGATACCAACGTCGGCAACAGGTGCCCAATGGATATATCGAGCCGCAACATGCTACCATATACTCAAGTTATCTCAGCCAGGGCGGTTACGGCGCTCGGCAGTACGGTTACGCTGGTAGAGCTGGTCCGTTCCTGCCGGGCTCGCTTCTCGGCTTCCAGAACGCGGCAGTCAACGAGCAGGCCCGCGCCATGCTCGGAGTCGCTGACATCAACTGGAAGGTCGGCGATCGCTGTCTAGCGTTATATTGGGAAGACAATAATTTCTACGAAGCGGAAATAACGGGCGTCTCCGCTAATACAGTTGTAGTTAAGTTCTGTGCATATGGTAATCACGAAGAAGTACTGAAATCCAACTGTCTACCATACCCTAATCAACCGGCGAGTGTGTAG
Protein
MSLRETLKELGWHLTEEGIDTITENGQIQDVNILSKKALDYDLRDISEGSLPEDFVKDPSKLESPVVVQIQKIRNVSAPKANEESTSAPRMLKLTLHDGKVTCTGLEISHIPSLSINTPPGTKLLLKNEELEVCHGVVWLTPSVISVLGGTVSHMIEKWELNRSLAKHTRGGIGAEGGPPPWIPFGQRLEALSVDKQFKSLQEAAKQDNPEFEAQRKGAIAEAQRMSGVKKVFGGGTKPLLDANVQKIVDAGFTEEQAEATLKYTKNNVDRALKILQKRDNSENRNKEKPQKEQDPVKRKGRHKESGDEDGVPVKPSGKVSLFDFLEDKLPNVPEKDKNTRQNNSSMEDRYDRSQNRERNSNRNRHGSRHEGANRNRSDRRHYSNDNLYSSYREDRKYQSQNEKPPRFQKIQEEMNKHHQNMNMPYNNTYQQNQYKDRHMRNDRNPNDRGPQQNYNHNSSTMDGLVDATSKLNLLSNQSRIDEQQVHNYQQHTDQSYQSKQTYPSNHSQDIPPFRRSNDLHQDGYQRRQQVPNGYIEPQHATIYSSYLSQGGYGARQYGYAGRAGPFLPGSLLGFQNAAVNEQARAMLGVADINWKVGDRCLALYWEDNNFYEAEITGVSANTVVVKFCAYGNHEEVLKSNCLPYPNQPASV
Summary
Description
Scaffolding protein that specifically recognizes and binds dimethylarginine-containing proteins. In nucleus, acts as a coactivator: recognizes and binds asymmetric dimethylation on the core histone tails associated with transcriptional activation (H3R17me2a and H4R3me2a) and recruits proteins at these arginine-methylated loci. In cytoplasm, may play a role in the assembly and/or disassembly of mRNA stress granules and in the regulation of translation of target mRNAs by binding Arg/Gly-rich motifs (GAR) in dimethylarginine-containing proteins (By similarity).
Subunit
Component of mRNA stress granules.
Keywords
Chromatin regulator
Complete proteome
Cytoplasm
Nucleus
Reference proteome
Feature
chain Tudor domain-containing protein 3
Uniprot
EMBL
BABH01000410
BABH01000411
AGBW02011847
OWR46030.1
NWSH01001158
PCG72313.1
+ More
KQ459439 KPJ01004.1 ODYU01000175 SOQ34517.1 GBGD01000762 JAC88127.1 GFTR01007640 JAW08786.1 GECL01001964 JAP04160.1 KQ981685 KYN37997.1 GL451627 EFN78875.1 KQ977720 KYN00436.1 LBMM01000178 KMR04597.1 GECU01033400 JAS74306.1 GEDC01011601 JAS25697.1 KZ288192 PBC34344.1 KQ434827 KZC07660.1 KQ978747 KYN28683.1 KQ435794 KOX73599.1 KQ414666 KOC64944.1 AJ719308
KQ459439 KPJ01004.1 ODYU01000175 SOQ34517.1 GBGD01000762 JAC88127.1 GFTR01007640 JAW08786.1 GECL01001964 JAP04160.1 KQ981685 KYN37997.1 GL451627 EFN78875.1 KQ977720 KYN00436.1 LBMM01000178 KMR04597.1 GECU01033400 JAS74306.1 GEDC01011601 JAS25697.1 KZ288192 PBC34344.1 KQ434827 KZC07660.1 KQ978747 KYN28683.1 KQ435794 KOX73599.1 KQ414666 KOC64944.1 AJ719308
Proteomes
Interpro
IPR009060
UBA-like_sf
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR013894 RMI1_N
IPR017986 WD40_repeat_dom
IPR010304 Survival_motor_neuron
IPR024977 Apc4_WD40_dom
IPR015940 UBA
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR002999 Tudor
IPR019775 WD40_repeat_CS
IPR033472 DUF1767
IPR041915 UBA_TDRD3
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR013894 RMI1_N
IPR017986 WD40_repeat_dom
IPR010304 Survival_motor_neuron
IPR024977 Apc4_WD40_dom
IPR015940 UBA
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR002999 Tudor
IPR019775 WD40_repeat_CS
IPR033472 DUF1767
IPR041915 UBA_TDRD3
Gene 3D
ProteinModelPortal
PDB
5GVE
E-value=3.91937e-32,
Score=347
Ontologies
KEGG
GO
Topology
Subcellular location
Cytoplasm
Predominantly cytoplasmic. Associated with actively translating polyribosomes and with mRNA stress granules (By similarity). With evidence from 1 publications.
Nucleus Predominantly cytoplasmic. Associated with actively translating polyribosomes and with mRNA stress granules (By similarity). With evidence from 1 publications.
Nucleus Predominantly cytoplasmic. Associated with actively translating polyribosomes and with mRNA stress granules (By similarity). With evidence from 1 publications.
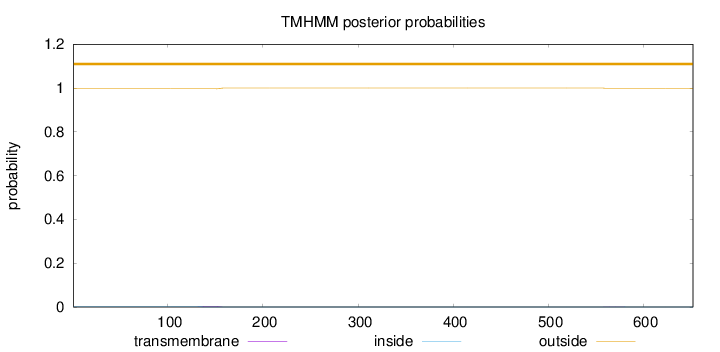
Length:
652
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08181
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00363
outside
1 - 652
Population Genetic Test Statistics
Pi
19.808725
Theta
15.949258
Tajima's D
0.329347
CLR
0.678494
CSRT
0.471126443677816
Interpretation
Uncertain
