Pre Gene Modal
BGIBMGA001123
Annotation
PREDICTED:_exportin-5_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 1.179
Sequence
CDS
ATGATGGCGGCCCCACTAGTGACGTGGTCGTACGTGGAGCAGTGGACGCAGTCGGCGCTGGACAAGCTGGAGTCGTGTCCGCGCGCGCTCCACACGCAGCACCCGCTCTACGTCGAGTGGGAGGCGCTCACGCAGGCGCTGGACGTGGTGCTGTCGCGGCTGGTGGAGACGGAGCCCCGGCCCAGCGCCGCGGCCGGCCTGGCGCTGCTGCAGCGCTGCGTGCTCACACAGCCCTCCGACCCGCTCTTGCTGTCACTGTTATTGTCACTCATCTCCGCGCTCTTCGTGTTCCTCAGCTGCGCCTACAGCCAGCTAGCAGGTCCCGGGGTAGGGGCGGCGGGGGCAGAGCTGCTGCCGCGCGTGTTGGACAAGATATTCTCGTCGCTGGTGTACGAGGGCGCCGCCCCGGACCAGCGCCGCTCCCGCGCCGTCAAGAACGTGCGCCGCCACGCCGCCGGCCTGCTCGTCAAACTCGGCTCCAAGTACCCGCTGTTACTGCTGCCGGTGTTCGGTCGCCTGCACGAGCTGTCCCGCGCGGCGCTGGCGCGGCCCGACCTCAGCGTAGTGGAGAGCGTGACGCTGCAGGAGGCGCTGCTGCTAGTCTCCAATCACTTCTGTTGCTACGAGAGGCAGAGCGCACTCGTCGCGCAGGTGGTCGGCGAATGCGGTCCGCGCTGGGCAGCGCTGAGCTCGCAGGCGGGCTCCGCGACGGCTCTGGTGCGCCTCATAGGGCTCGACGCGGCGCCCGTGGACAGCTCGCCGAGCGACACGGACGGGCCCGGCTCGGCGCGGCGCGACCTGCTGCACGGCCTCACGCTGCTGCTGGGCGTCGTGAAGCGCACGCACGTGCCCGCCGACCCCGACAAGGCCACGCGCGGCGGGTTCAGCGCCGGCCTCACCGCCGCCGGCAACCCCGTGTGGCGCAACCCCTGCGCCGCGCACGTGCTGCCGCTGTTCCCCGGCGCGCTGGCGCTGGGCCGCGCGCTGCACGAGCTGCACGCGGCGGGCGCGGCGGGGCTGCTGCACGCGGGGCAGGCGCGCGCGCTGGGCCCGCCCGCCGCCGAGCGCCGCAACCTGCTGGGCCTGTGCGACCAGGACGCGGCGCCCGTGTCCCCGGCGTCGCGCATGCAGCAGTACGTGCACACGCTGCACGAGTCCGTGTGCCACCTGGTGGGCGCGGCGGCGGCGTCGCTGGGCCGCGAGCTGTACGCGGCGCCGGGGCTGGCGCGGGCGCTGGCGGGCTCGCTGCTGCAGGGGCTGGACCGGCTGCCGGACCACTGGCTGCGGCCGCTCGTCCGATGCGCGCTCAAGCCTCTCATCTTGCACTGTCCGCCAGCCCACTATCACGACGTCGCCGTACCAATACTCGAACACGTGGCCCCGTTCATGTTAATGCATCTGTCGCAACGATGGGACTACATCATGTCTCTCTACGAGTCTGGTAAACTAGAAGAGGAAACCGGCAGTGAGTCTCAAGAAGTGTTAGAAGATATCCTCGTTAGAAACCTGACTAAAGAATATTTAGAAGTTATTAAAGTGTCGCTGGTGGAGGGCGGCGTGGTGTCAAGTGCGGACACGTGCGACGACATGGAGGAGGACTCCAGCCCCGCGCCCCCCGCGCCCGCCGCCGCGCACACCCGCACGCCCGACGCCGTGTCCGAGCTGGGCGCGCTGCTGCTGCAGCATCCGGTCGCGGGACCCGCTCTCTTGCACACGGTACTGCGTGCGCTGACGTGGGGCGACTCGAGCGCGTCGCTGCGCGCGTGCGGGCTGGCGCTGGCGGCGGTGGCGGGCGCGCGCGCGGCGGGGCAGCTGGGCGCGGCGGAGGCGGCGGGCGCGCTGCACGCCCAGCTGCACGCCCTGCGCGCGCACGGACAGCACGACGCCAACCAGGCCGCACTGCTCGCGCTCGCGCTGCAGACATACGAAACTCTCCGGCCACAGTTCCCTAACATAGTAAATGTACTACGTGCCATTCCGGATGTGGACGCACACGATCTGCAGCGGCTAGAAGATAAGCTTGCCAACAACACCGGGAAACCTTCCAAAATCGACAAGAGCAAACGAGATTTATTTAAGAAAATCACTTCCAAGTTAATTGGTCGCAACGTGGGTCAGCTGTTCAAGAAGGAAGTGTTCATAGTGGATCTGCCCACGGTCCGCACGGCTCGCCCGCCGACTGACGACACCGCCCCGCCCCGACTGGACCTGCTGTTCGCGCACACTCCACCCACCTAG
Protein
MMAAPLVTWSYVEQWTQSALDKLESCPRALHTQHPLYVEWEALTQALDVVLSRLVETEPRPSAAAGLALLQRCVLTQPSDPLLLSLLLSLISALFVFLSCAYSQLAGPGVGAAGAELLPRVLDKIFSSLVYEGAAPDQRRSRAVKNVRRHAAGLLVKLGSKYPLLLLPVFGRLHELSRAALARPDLSVVESVTLQEALLLVSNHFCCYERQSALVAQVVGECGPRWAALSSQAGSATALVRLIGLDAAPVDSSPSDTDGPGSARRDLLHGLTLLLGVVKRTHVPADPDKATRGGFSAGLTAAGNPVWRNPCAAHVLPLFPGALALGRALHELHAAGAAGLLHAGQARALGPPAAERRNLLGLCDQDAAPVSPASRMQQYVHTLHESVCHLVGAAAASLGRELYAAPGLARALAGSLLQGLDRLPDHWLRPLVRCALKPLILHCPPAHYHDVAVPILEHVAPFMLMHLSQRWDYIMSLYESGKLEEETGSESQEVLEDILVRNLTKEYLEVIKVSLVEGGVVSSADTCDDMEEDSSPAPPAPAAAHTRTPDAVSELGALLLQHPVAGPALLHTVLRALTWGDSSASLRACGLALAAVAGARAAGQLGAAEAAGALHAQLHALRAHGQHDANQAALLALALQTYETLRPQFPNIVNVLRAIPDVDAHDLQRLEDKLANNTGKPSKIDKSKRDLFKKITSKLIGRNVGQLFKKEVFIVDLPTVRTARPPTDDTAPPRLDLLFAHTPPT
Summary
Uniprot
A0A2H1V186
A0A194QC41
A0A212EX50
A0A194RCW0
A0A2H1WS44
A0A2A3EBT3
+ More
A0A088A7B5 E9IV21 A0A310SI85 A0A0N0BCA4 A0A0L7R2Y9 A0A026X2H1 E2AU91 A0A151X223 A0A151IQV0 A0A0J7KYW3 A0A1L8DXG8 A0A182WDY0 A0A084WIN9 E2B373 A0A195AVD8 A0A195E423 A0A158P273 A0A182RRZ8 A0A232F2R4 F4W6G2 A0A182UKU0 A0A182UXB6 U5EYH2 A0A182MSS8 A0A182PKE2 A0A182Y9F3 A0A182L513 A0A182HHI4 Q7PZF9 A0A182JRB2 A0A182XJV0 A0A182SNG1 A0A182IRS2 A0A182QTA7 A0A182FB47 A0A195FI36 A0A154PPI6 A0A0C9PKR9 A0A182N2W6 D2A2P3 A0A0T6B8N7 A0A1B0D7F5 B0WTT9 A0A0A1WJW8 A0A1Q3EYL6 A0A0K8V1J6 A0A1Q3EYK0 A0A1Q3EYH6 A0A1Q3EYG9 A0A1W4XC73 W5JX12 A0A336LWS5 A0A1Y1M205 Q16VX2 A0A1I8NAX9 A0A1S4FM74 A0A067R4D7 A0A2J7Q556 A0A1I8PEX8 A0A1A9WDP4 A0A0L0CFM5 A0A1L8DXD8 A0A182GAX7 A0A182G252 A0A1A9VRC2 A0A1B0BWU4 A0A1A9XCU1 A0A1B0A058 A0A1B0BB62 A0A1B0FME9 A0A3B0JA48 T1GUQ9 Q29IU1 B4L7Y9 N6UU42 U4TWW1 A0A0V0GA69 B4MAQ2 B4JK81 Q2XXY8 A0A146KWR0 Q9BP41 Q9VWE7 A0A1J1IPB8 B4MSQ4 Q2XXZ0 Q2XXY6 A0A1W4VSZ3 A0A1W4VG06 Q2XXY9 B4PZE6 B3NVR7 Q2XXY5
A0A088A7B5 E9IV21 A0A310SI85 A0A0N0BCA4 A0A0L7R2Y9 A0A026X2H1 E2AU91 A0A151X223 A0A151IQV0 A0A0J7KYW3 A0A1L8DXG8 A0A182WDY0 A0A084WIN9 E2B373 A0A195AVD8 A0A195E423 A0A158P273 A0A182RRZ8 A0A232F2R4 F4W6G2 A0A182UKU0 A0A182UXB6 U5EYH2 A0A182MSS8 A0A182PKE2 A0A182Y9F3 A0A182L513 A0A182HHI4 Q7PZF9 A0A182JRB2 A0A182XJV0 A0A182SNG1 A0A182IRS2 A0A182QTA7 A0A182FB47 A0A195FI36 A0A154PPI6 A0A0C9PKR9 A0A182N2W6 D2A2P3 A0A0T6B8N7 A0A1B0D7F5 B0WTT9 A0A0A1WJW8 A0A1Q3EYL6 A0A0K8V1J6 A0A1Q3EYK0 A0A1Q3EYH6 A0A1Q3EYG9 A0A1W4XC73 W5JX12 A0A336LWS5 A0A1Y1M205 Q16VX2 A0A1I8NAX9 A0A1S4FM74 A0A067R4D7 A0A2J7Q556 A0A1I8PEX8 A0A1A9WDP4 A0A0L0CFM5 A0A1L8DXD8 A0A182GAX7 A0A182G252 A0A1A9VRC2 A0A1B0BWU4 A0A1A9XCU1 A0A1B0A058 A0A1B0BB62 A0A1B0FME9 A0A3B0JA48 T1GUQ9 Q29IU1 B4L7Y9 N6UU42 U4TWW1 A0A0V0GA69 B4MAQ2 B4JK81 Q2XXY8 A0A146KWR0 Q9BP41 Q9VWE7 A0A1J1IPB8 B4MSQ4 Q2XXZ0 Q2XXY6 A0A1W4VSZ3 A0A1W4VG06 Q2XXY9 B4PZE6 B3NVR7 Q2XXY5
Pubmed
26354079
22118469
21282665
24508170
20798317
24438588
+ More
21347285 28648823 21719571 25244985 20966253 12364791 14747013 17210077 18362917 19820115 25830018 20920257 23761445 28004739 17510324 25315136 24845553 26108605 26483478 15632085 17994087 18057021 23537049 16120803 26823975 12426392 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
21347285 28648823 21719571 25244985 20966253 12364791 14747013 17210077 18362917 19820115 25830018 20920257 23761445 28004739 17510324 25315136 24845553 26108605 26483478 15632085 17994087 18057021 23537049 16120803 26823975 12426392 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
ODYU01000175
SOQ34516.1
KQ459439
KPJ01001.1
AGBW02011847
OWR46027.1
+ More
KQ460397 KPJ15304.1 ODYU01010583 SOQ55796.1 KZ288296 PBC28934.1 GL766116 EFZ15568.1 KQ759878 OAD62258.1 KQ435918 KOX68731.1 KQ414664 KOC65194.1 KK107054 EZA61584.1 GL442815 EFN62948.1 KQ982580 KYQ54462.1 KQ976755 KYN08593.1 LBMM01001958 KMQ95474.1 GFDF01002953 JAV11131.1 ATLV01023941 KE525347 KFB50083.1 GL445305 EFN89855.1 KQ976731 KYM76198.1 KQ979657 KYN19903.1 ADTU01007003 NNAY01001104 OXU25116.1 GL887707 EGI70297.1 GANO01000360 JAB59511.1 AXCM01009764 APCN01002416 AAAB01008986 EAA00125.3 AXCN02001090 KQ981523 KYN40350.1 KQ435012 KZC13806.1 GBYB01001603 JAG71370.1 KQ971338 EFA02755.2 LJIG01009125 KRT83706.1 AJVK01003920 DS232092 EDS34630.1 GBXI01014978 JAC99313.1 GFDL01014665 JAV20380.1 GDHF01027080 GDHF01019839 GDHF01012830 JAI25234.1 JAI32475.1 JAI39484.1 GFDL01014660 JAV20385.1 GFDL01014690 JAV20355.1 GFDL01014691 JAV20354.1 ADMH02000120 ETN67749.1 UFQT01000174 SSX21113.1 GEZM01042911 GEZM01042910 JAV79481.1 CH477579 EAT38741.1 KK852760 KDR17037.1 NEVH01018372 PNF23710.1 JRES01000455 KNC31045.1 GFDF01002957 JAV11127.1 JXUM01051680 JXUM01051681 KQ561700 KXJ77769.1 JXUM01039442 JXUM01039443 JXUM01039444 KQ561204 KXJ79361.1 JXJN01021970 JXJN01011306 CCAG010005910 OUUW01000003 SPP78835.1 CAQQ02129626 CAQQ02129627 CAQQ02129628 CH379063 EAL32562.1 CH933814 EDW05564.1 KRG07451.1 KRG07452.1 APGK01014224 KB739238 ENN82327.1 KB631581 ERL84473.1 GECL01001169 JAP04955.1 CH940655 EDW66311.1 KRF82632.1 CH916370 EDV99983.1 DQ138874 ABA86480.1 GDHC01017951 JAQ00678.1 AF222746 AAG53520.1 AE014298 AY118535 AAF48995.1 AAM49904.1 AHN59923.1 CVRI01000055 CRL01396.1 CH963851 EDW75143.1 DQ138872 ABA86478.1 DQ138875 DQ138876 ABA86481.1 ABA86482.1 DQ138873 ABA86479.1 CM000162 EDX02102.1 CH954180 EDV46732.1 DQ138877 ABA86483.1
KQ460397 KPJ15304.1 ODYU01010583 SOQ55796.1 KZ288296 PBC28934.1 GL766116 EFZ15568.1 KQ759878 OAD62258.1 KQ435918 KOX68731.1 KQ414664 KOC65194.1 KK107054 EZA61584.1 GL442815 EFN62948.1 KQ982580 KYQ54462.1 KQ976755 KYN08593.1 LBMM01001958 KMQ95474.1 GFDF01002953 JAV11131.1 ATLV01023941 KE525347 KFB50083.1 GL445305 EFN89855.1 KQ976731 KYM76198.1 KQ979657 KYN19903.1 ADTU01007003 NNAY01001104 OXU25116.1 GL887707 EGI70297.1 GANO01000360 JAB59511.1 AXCM01009764 APCN01002416 AAAB01008986 EAA00125.3 AXCN02001090 KQ981523 KYN40350.1 KQ435012 KZC13806.1 GBYB01001603 JAG71370.1 KQ971338 EFA02755.2 LJIG01009125 KRT83706.1 AJVK01003920 DS232092 EDS34630.1 GBXI01014978 JAC99313.1 GFDL01014665 JAV20380.1 GDHF01027080 GDHF01019839 GDHF01012830 JAI25234.1 JAI32475.1 JAI39484.1 GFDL01014660 JAV20385.1 GFDL01014690 JAV20355.1 GFDL01014691 JAV20354.1 ADMH02000120 ETN67749.1 UFQT01000174 SSX21113.1 GEZM01042911 GEZM01042910 JAV79481.1 CH477579 EAT38741.1 KK852760 KDR17037.1 NEVH01018372 PNF23710.1 JRES01000455 KNC31045.1 GFDF01002957 JAV11127.1 JXUM01051680 JXUM01051681 KQ561700 KXJ77769.1 JXUM01039442 JXUM01039443 JXUM01039444 KQ561204 KXJ79361.1 JXJN01021970 JXJN01011306 CCAG010005910 OUUW01000003 SPP78835.1 CAQQ02129626 CAQQ02129627 CAQQ02129628 CH379063 EAL32562.1 CH933814 EDW05564.1 KRG07451.1 KRG07452.1 APGK01014224 KB739238 ENN82327.1 KB631581 ERL84473.1 GECL01001169 JAP04955.1 CH940655 EDW66311.1 KRF82632.1 CH916370 EDV99983.1 DQ138874 ABA86480.1 GDHC01017951 JAQ00678.1 AF222746 AAG53520.1 AE014298 AY118535 AAF48995.1 AAM49904.1 AHN59923.1 CVRI01000055 CRL01396.1 CH963851 EDW75143.1 DQ138872 ABA86478.1 DQ138875 DQ138876 ABA86481.1 ABA86482.1 DQ138873 ABA86479.1 CM000162 EDX02102.1 CH954180 EDV46732.1 DQ138877 ABA86483.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000242457
UP000005203
UP000053105
+ More
UP000053825 UP000053097 UP000000311 UP000075809 UP000078542 UP000036403 UP000075920 UP000030765 UP000008237 UP000078540 UP000078492 UP000005205 UP000075900 UP000215335 UP000007755 UP000075902 UP000075903 UP000075883 UP000075885 UP000076408 UP000075882 UP000075840 UP000007062 UP000075881 UP000076407 UP000075901 UP000075880 UP000075886 UP000069272 UP000078541 UP000076502 UP000075884 UP000007266 UP000092462 UP000002320 UP000192223 UP000000673 UP000008820 UP000095301 UP000027135 UP000235965 UP000095300 UP000091820 UP000037069 UP000069940 UP000249989 UP000078200 UP000092460 UP000092443 UP000092445 UP000092444 UP000268350 UP000015102 UP000001819 UP000009192 UP000019118 UP000030742 UP000008792 UP000001070 UP000000803 UP000183832 UP000007798 UP000192221 UP000002282 UP000008711
UP000053825 UP000053097 UP000000311 UP000075809 UP000078542 UP000036403 UP000075920 UP000030765 UP000008237 UP000078540 UP000078492 UP000005205 UP000075900 UP000215335 UP000007755 UP000075902 UP000075903 UP000075883 UP000075885 UP000076408 UP000075882 UP000075840 UP000007062 UP000075881 UP000076407 UP000075901 UP000075880 UP000075886 UP000069272 UP000078541 UP000076502 UP000075884 UP000007266 UP000092462 UP000002320 UP000192223 UP000000673 UP000008820 UP000095301 UP000027135 UP000235965 UP000095300 UP000091820 UP000037069 UP000069940 UP000249989 UP000078200 UP000092460 UP000092443 UP000092445 UP000092444 UP000268350 UP000015102 UP000001819 UP000009192 UP000019118 UP000030742 UP000008792 UP000001070 UP000000803 UP000183832 UP000007798 UP000192221 UP000002282 UP000008711
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2H1V186
A0A194QC41
A0A212EX50
A0A194RCW0
A0A2H1WS44
A0A2A3EBT3
+ More
A0A088A7B5 E9IV21 A0A310SI85 A0A0N0BCA4 A0A0L7R2Y9 A0A026X2H1 E2AU91 A0A151X223 A0A151IQV0 A0A0J7KYW3 A0A1L8DXG8 A0A182WDY0 A0A084WIN9 E2B373 A0A195AVD8 A0A195E423 A0A158P273 A0A182RRZ8 A0A232F2R4 F4W6G2 A0A182UKU0 A0A182UXB6 U5EYH2 A0A182MSS8 A0A182PKE2 A0A182Y9F3 A0A182L513 A0A182HHI4 Q7PZF9 A0A182JRB2 A0A182XJV0 A0A182SNG1 A0A182IRS2 A0A182QTA7 A0A182FB47 A0A195FI36 A0A154PPI6 A0A0C9PKR9 A0A182N2W6 D2A2P3 A0A0T6B8N7 A0A1B0D7F5 B0WTT9 A0A0A1WJW8 A0A1Q3EYL6 A0A0K8V1J6 A0A1Q3EYK0 A0A1Q3EYH6 A0A1Q3EYG9 A0A1W4XC73 W5JX12 A0A336LWS5 A0A1Y1M205 Q16VX2 A0A1I8NAX9 A0A1S4FM74 A0A067R4D7 A0A2J7Q556 A0A1I8PEX8 A0A1A9WDP4 A0A0L0CFM5 A0A1L8DXD8 A0A182GAX7 A0A182G252 A0A1A9VRC2 A0A1B0BWU4 A0A1A9XCU1 A0A1B0A058 A0A1B0BB62 A0A1B0FME9 A0A3B0JA48 T1GUQ9 Q29IU1 B4L7Y9 N6UU42 U4TWW1 A0A0V0GA69 B4MAQ2 B4JK81 Q2XXY8 A0A146KWR0 Q9BP41 Q9VWE7 A0A1J1IPB8 B4MSQ4 Q2XXZ0 Q2XXY6 A0A1W4VSZ3 A0A1W4VG06 Q2XXY9 B4PZE6 B3NVR7 Q2XXY5
A0A088A7B5 E9IV21 A0A310SI85 A0A0N0BCA4 A0A0L7R2Y9 A0A026X2H1 E2AU91 A0A151X223 A0A151IQV0 A0A0J7KYW3 A0A1L8DXG8 A0A182WDY0 A0A084WIN9 E2B373 A0A195AVD8 A0A195E423 A0A158P273 A0A182RRZ8 A0A232F2R4 F4W6G2 A0A182UKU0 A0A182UXB6 U5EYH2 A0A182MSS8 A0A182PKE2 A0A182Y9F3 A0A182L513 A0A182HHI4 Q7PZF9 A0A182JRB2 A0A182XJV0 A0A182SNG1 A0A182IRS2 A0A182QTA7 A0A182FB47 A0A195FI36 A0A154PPI6 A0A0C9PKR9 A0A182N2W6 D2A2P3 A0A0T6B8N7 A0A1B0D7F5 B0WTT9 A0A0A1WJW8 A0A1Q3EYL6 A0A0K8V1J6 A0A1Q3EYK0 A0A1Q3EYH6 A0A1Q3EYG9 A0A1W4XC73 W5JX12 A0A336LWS5 A0A1Y1M205 Q16VX2 A0A1I8NAX9 A0A1S4FM74 A0A067R4D7 A0A2J7Q556 A0A1I8PEX8 A0A1A9WDP4 A0A0L0CFM5 A0A1L8DXD8 A0A182GAX7 A0A182G252 A0A1A9VRC2 A0A1B0BWU4 A0A1A9XCU1 A0A1B0A058 A0A1B0BB62 A0A1B0FME9 A0A3B0JA48 T1GUQ9 Q29IU1 B4L7Y9 N6UU42 U4TWW1 A0A0V0GA69 B4MAQ2 B4JK81 Q2XXY8 A0A146KWR0 Q9BP41 Q9VWE7 A0A1J1IPB8 B4MSQ4 Q2XXZ0 Q2XXY6 A0A1W4VSZ3 A0A1W4VG06 Q2XXY9 B4PZE6 B3NVR7 Q2XXY5
PDB
5YU7
E-value=1.10984e-69,
Score=672
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
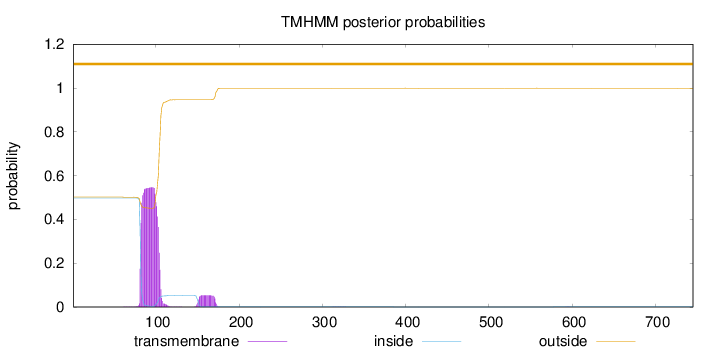
Length:
745
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.1939899999999
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.49805
outside
1 - 745
Population Genetic Test Statistics
Pi
24.091015
Theta
23.119546
Tajima's D
-1.355774
CLR
1.494252
CSRT
0.0791960401979901
Interpretation
Uncertain
