Gene
KWMTBOMO08040 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001035
Annotation
PREDICTED:_inosine-5'-monophosphate_dehydrogenase_[Amyelois_transitella]
Full name
Inosine-5'-monophosphate dehydrogenase
Alternative Name
Protein raspberry
Location in the cell
Cytoplasmic Reliability : 1.609 Mitochondrial Reliability : 1.583
Sequence
CDS
ATGTCATTAAAGACGGAAGGCGACCTCCGAGATGGGTTATCTGCTGAGGATACCTTCGCTAACAGTGAAGGACTTACATATAACGATTTCTTACTGCTGCCTGGATATATAGATTTTACAGCAGAAGAAGTCGATTTGACATCTCCACTCACTAAGAAAATTCTGTTAAAAGCACCTCTAGTGTCTACTCCAATGGACACAGTGACAGAAGCTGACATGGCCATATCCATGGCGCTTTGCGGTGGAATTGGCATTATACACCACAACTGTACTCCAGAATACCAAGCTAATGAAGTTCACAAAGTGAAGAAGTATAAGCATGGATTTATTAGAGACCCCGTTTGCATGGGTCCTGAGAACACAGTCGCTGATGTCTTGGAAGCTAAGAAGAAGAACGGATTCACTGGCTATCCTATCACTCAGAATGGTAAACTTGGTGGACGTCTAATTGGCATTGTTACATCTCGTGACATTGATTTTAGAGAAGGGGACCCACACCTAAGTTTGAAAGAAGTAATGACTCCCATAAATGATATGATTACTGCACAATTGGGTGTCACTTTACAAGATGCTAATTACATATTAGAGAAGAGCAAAAAAGGAAAATTACCAATCATTAATAATGATGGTGAACTTGTTGCATTGATTGCTAGAACTGATTTAAAGAAAGCGAGAAGTTATCCAAATGCATCTAAAGATTCCAACAAGCAATTGTTAGTAGGGGCGGCAATTGGGACTCGTGATACAGACAGAGAACGGCTGAAGTTGCTTGTGAACAATGGTGTAGATGTGATTGTCCTCGATTCATCTCAGGGAAATTCAACCTACCAAATCAAGATGATAAAATACATAAAGGAAACTTATCCAGAAATACAGGTAGTTGGTGGTAATGTAGTGACAAGAATGCAAGCTAAAAATTTAATTGACGCAGGAGTTGATGCCTTGAGAGTCGGAATGGGTAGTGGCTCTATTTGTATCACACAAGAAGTGATGGCTTGTGGTTGTCCTCAAGCCACTGCAGTGTACCAAGTAGCTTCCTATGCTCGTCATTTTAATGTACCAGTCATTGCCGATGGTGGAATTCAGTCAGTTGGTCATATTATTAAATCATTGGCCCTTGGGGCATCGACTGTAATGATGGGATCACTCCTTGCGGGGACATCTGAGGCCCCTGGAGAATACTTCTTCTCTGATGGAGTTCGGCTCAAGAAATACAGAGGAATGGGAAGCTTAGAAGCTATGGAAAGTAAAGATGGTAAAGGATCTGCTATGAGTAGATACTTCCACAAAGAATCTGACAAACATAGAGTTGCTCAGGGAGTTAGTGGTAGTATTGTTGATAAAGGTTCAGTACTAAGATTCCTACCTTATTTGCAAGCCGGCATGCAACACAGTTGCCAAGATCTTGGAGCTCGATCTGTTAGTGTGTTGCGTGAAATGAGTCATTCTGGAGATCTGAGATTCATGAAAAGGACTTACTCAGCCCAACTGGAAGGCAATGTCCATGGTCTGTTCTCTTATGAGAAAAGATTATTTTAA
Protein
MSLKTEGDLRDGLSAEDTFANSEGLTYNDFLLLPGYIDFTAEEVDLTSPLTKKILLKAPLVSTPMDTVTEADMAISMALCGGIGIIHHNCTPEYQANEVHKVKKYKHGFIRDPVCMGPENTVADVLEAKKKNGFTGYPITQNGKLGGRLIGIVTSRDIDFREGDPHLSLKEVMTPINDMITAQLGVTLQDANYILEKSKKGKLPIINNDGELVALIARTDLKKARSYPNASKDSNKQLLVGAAIGTRDTDRERLKLLVNNGVDVIVLDSSQGNSTYQIKMIKYIKETYPEIQVVGGNVVTRMQAKNLIDAGVDALRVGMGSGSICITQEVMACGCPQATAVYQVASYARHFNVPVIADGGIQSVGHIIKSLALGASTVMMGSLLAGTSEAPGEYFFSDGVRLKKYRGMGSLEAMESKDGKGSAMSRYFHKESDKHRVAQGVSGSIVDKGSVLRFLPYLQAGMQHSCQDLGARSVSVLREMSHSGDLRFMKRTYSAQLEGNVHGLFSYEKRLF
Summary
Description
Catalyzes the conversion of inosine 5'-phosphate (IMP) to xanthosine 5'-phosphate (XMP), the first committed and rate-limiting step in the de novo synthesis of guanine nucleotides, and therefore plays an important role in the regulation of cell growth.
Catalytic Activity
H2O + IMP + NAD(+) = H(+) + NADH + XMP
Cofactor
K(+)
Subunit
Homotetramer.
Similarity
Belongs to the IMPDH/GMPR family.
Keywords
CBS domain
Complete proteome
Cytoplasm
GMP biosynthesis
Metal-binding
NAD
Oxidoreductase
Phosphoprotein
Potassium
Purine biosynthesis
Reference proteome
Repeat
Feature
chain Inosine-5'-monophosphate dehydrogenase
Uniprot
H9IUV5
A0A2H1UZU7
A0A0N0PEW2
A0A194RCV6
S4PG58
A0A0L7LSA9
+ More
Q16WB3 A0A182NZP6 A0A336KX26 A0A182X5P8 Q7QHD0 A0A1S4H5T8 A0A182TPA4 A0A182HW99 A0A182KL07 A0A023EUH8 A0A182JWZ7 A0A182UVK4 A0A182N290 A0A182RW44 A0A182YB03 A0A0K8TPP4 A0A182SCF6 W8BA97 A0A0A1XHQ0 W5JN05 A0A034W0T2 A0A0K8VZ52 A0A2M4AB86 A0A182Q952 A0A182FW38 A0A182MPN7 A0A194Q7M2 A0A1Q3EWZ0 A0A1J1J3M4 A0A2M3Z269 A0A2M4BKR2 A0A182WDL1 B0WW86 A0A1I8QE28 A4V488 Q07152 B4PXZ1 T1P9Q0 B4MJ75 B3MSB7 A0A3B0JYW0 A0A1W4UNW8 A0A0L0C2R1 B3NW07 Q29IQ0 A0A182IPX8 B4MES0 A0A1A9ZRR6 B4JK62 A0A0M3QZA3 U5EIP0 A0A1A9WCQ4 A0A1B0ATC8 A0A1A9XAI2 A0A1A9UPG8 A0A1A9VPM0 A0A1A9WDG8 A0A1B0FR56 A0A1A9YA98 A0A1A9ZJM4 B4R7M3 B4IDT3 A0A1B0GGT8 B4H2U5 A0A1L8DXU5 A0A084WM01 A0A067RHP6 A0A0C9R572 E2BHJ7 A0A154P3W3 A0A2P8XQG3 A0A1Y1JWM5 A0A0M9A7J8 A0A2J7Q0K4 A0A232F3Y3 A0A0P4VV19 A0A2A3EIS3 A0A088A653 A0A0L7R1T3 A0A2R7W2W8 A0A151I6G9 A0A151I6F4 A0A0V0G8A7 A0A224XAV9 A0A069DUB9 A0A1Y1JWM0 A0A1W4WQG2 A0A151IUY9 E2A771 A0A151K3Z5 F4WE14 A0A195BAH6 A0A158NUS4 A0A1B6EDT5
Q16WB3 A0A182NZP6 A0A336KX26 A0A182X5P8 Q7QHD0 A0A1S4H5T8 A0A182TPA4 A0A182HW99 A0A182KL07 A0A023EUH8 A0A182JWZ7 A0A182UVK4 A0A182N290 A0A182RW44 A0A182YB03 A0A0K8TPP4 A0A182SCF6 W8BA97 A0A0A1XHQ0 W5JN05 A0A034W0T2 A0A0K8VZ52 A0A2M4AB86 A0A182Q952 A0A182FW38 A0A182MPN7 A0A194Q7M2 A0A1Q3EWZ0 A0A1J1J3M4 A0A2M3Z269 A0A2M4BKR2 A0A182WDL1 B0WW86 A0A1I8QE28 A4V488 Q07152 B4PXZ1 T1P9Q0 B4MJ75 B3MSB7 A0A3B0JYW0 A0A1W4UNW8 A0A0L0C2R1 B3NW07 Q29IQ0 A0A182IPX8 B4MES0 A0A1A9ZRR6 B4JK62 A0A0M3QZA3 U5EIP0 A0A1A9WCQ4 A0A1B0ATC8 A0A1A9XAI2 A0A1A9UPG8 A0A1A9VPM0 A0A1A9WDG8 A0A1B0FR56 A0A1A9YA98 A0A1A9ZJM4 B4R7M3 B4IDT3 A0A1B0GGT8 B4H2U5 A0A1L8DXU5 A0A084WM01 A0A067RHP6 A0A0C9R572 E2BHJ7 A0A154P3W3 A0A2P8XQG3 A0A1Y1JWM5 A0A0M9A7J8 A0A2J7Q0K4 A0A232F3Y3 A0A0P4VV19 A0A2A3EIS3 A0A088A653 A0A0L7R1T3 A0A2R7W2W8 A0A151I6G9 A0A151I6F4 A0A0V0G8A7 A0A224XAV9 A0A069DUB9 A0A1Y1JWM0 A0A1W4WQG2 A0A151IUY9 E2A771 A0A151K3Z5 F4WE14 A0A195BAH6 A0A158NUS4 A0A1B6EDT5
EC Number
1.1.1.205
Pubmed
19121390
26354079
23622113
26227816
17510324
12364791
+ More
20966253 24945155 25244985 26369729 24495485 25830018 20920257 23761445 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7911114 7904480 7476879 12537569 18327897 17994087 17550304 25315136 26108605 15632085 23185243 24438588 24845553 20798317 29403074 28004739 28648823 27129103 26334808 21719571 21347285
20966253 24945155 25244985 26369729 24495485 25830018 20920257 23761445 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7911114 7904480 7476879 12537569 18327897 17994087 17550304 25315136 26108605 15632085 23185243 24438588 24845553 20798317 29403074 28004739 28648823 27129103 26334808 21719571 21347285
EMBL
BABH01000426
ODYU01000029
SOQ34111.1
LADI01003404
KPJ20584.1
KQ460397
+ More
KPJ15299.1 GAIX01003777 JAA88783.1 JTDY01000244 KOB78081.1 CH477571 EAT38872.1 UFQS01001062 UFQT01000002 UFQT01001062 SSX08781.1 SSX17088.1 AAAB01008816 EAA05291.2 APCN01001478 GAPW01001007 JAC12591.1 GDAI01001733 JAI15870.1 GAMC01012602 JAB93953.1 GBXI01003781 JAD10511.1 ADMH02000909 ETN64698.1 GAKP01010675 GAKP01010674 GAKP01010673 GAKP01010672 JAC48279.1 GDHF01008464 JAI43850.1 GGFK01004699 MBW38020.1 AXCN02001731 AXCM01004806 KQ459439 KPJ00995.1 GFDL01015221 JAV19824.1 CVRI01000064 CRL05489.1 GGFM01001871 MBW22622.1 GGFJ01004412 MBW53553.1 DS232140 EDS35918.1 AE014298 BT125958 AAN09265.1 ADX35937.1 L14847 L22608 S80430 AY089553 CM000162 EDX02963.1 KA645471 AFP60100.1 CH963738 EDW72164.1 CH902622 EDV34672.1 OUUW01000003 SPP78869.1 JRES01000982 KNC26522.1 CH954180 EDV46140.1 CH379063 EAL32603.2 CH940664 EDW63045.1 CH916370 EDV99964.1 CP012528 ALC49073.1 GANO01002555 JAB57316.1 JXJN01003238 CCAG010000549 CM000366 EDX17582.1 CH480830 EDW45741.1 AJWK01000737 AJWK01000738 CH479204 EDW30662.1 GFDF01002816 JAV11268.1 ATLV01024336 ATLV01024337 KE525351 KFB51245.1 KK852463 KDR23322.1 GBYB01007932 JAG77699.1 GL448301 EFN84850.1 KQ434809 KZC06512.1 PYGN01001531 PSN34238.1 GEZM01098856 GEZM01098855 GEZM01098854 GEZM01098852 GEZM01098851 GEZM01098848 JAV53712.1 KQ435719 KOX78850.1 NEVH01019967 PNF22099.1 NNAY01001108 OXU25107.1 GDKW01000711 JAI55884.1 KZ288229 PBC31703.1 KQ414667 KOC64822.1 KK854265 PTY13929.1 KQ978473 KYM93699.1 KYM93697.1 GECL01001804 JAP04320.1 GFTR01006899 JAW09527.1 GBGD01001161 JAC87728.1 GEZM01098853 GEZM01098850 GEZM01098849 JAV53714.1 KQ980926 KYN11389.1 GL437267 EFN70727.1 LKEZ01003766 KYN50721.1 GL888102 EGI67452.1 KQ976532 KYM81551.1 ADTU01026689 GEDC01001215 JAS36083.1
KPJ15299.1 GAIX01003777 JAA88783.1 JTDY01000244 KOB78081.1 CH477571 EAT38872.1 UFQS01001062 UFQT01000002 UFQT01001062 SSX08781.1 SSX17088.1 AAAB01008816 EAA05291.2 APCN01001478 GAPW01001007 JAC12591.1 GDAI01001733 JAI15870.1 GAMC01012602 JAB93953.1 GBXI01003781 JAD10511.1 ADMH02000909 ETN64698.1 GAKP01010675 GAKP01010674 GAKP01010673 GAKP01010672 JAC48279.1 GDHF01008464 JAI43850.1 GGFK01004699 MBW38020.1 AXCN02001731 AXCM01004806 KQ459439 KPJ00995.1 GFDL01015221 JAV19824.1 CVRI01000064 CRL05489.1 GGFM01001871 MBW22622.1 GGFJ01004412 MBW53553.1 DS232140 EDS35918.1 AE014298 BT125958 AAN09265.1 ADX35937.1 L14847 L22608 S80430 AY089553 CM000162 EDX02963.1 KA645471 AFP60100.1 CH963738 EDW72164.1 CH902622 EDV34672.1 OUUW01000003 SPP78869.1 JRES01000982 KNC26522.1 CH954180 EDV46140.1 CH379063 EAL32603.2 CH940664 EDW63045.1 CH916370 EDV99964.1 CP012528 ALC49073.1 GANO01002555 JAB57316.1 JXJN01003238 CCAG010000549 CM000366 EDX17582.1 CH480830 EDW45741.1 AJWK01000737 AJWK01000738 CH479204 EDW30662.1 GFDF01002816 JAV11268.1 ATLV01024336 ATLV01024337 KE525351 KFB51245.1 KK852463 KDR23322.1 GBYB01007932 JAG77699.1 GL448301 EFN84850.1 KQ434809 KZC06512.1 PYGN01001531 PSN34238.1 GEZM01098856 GEZM01098855 GEZM01098854 GEZM01098852 GEZM01098851 GEZM01098848 JAV53712.1 KQ435719 KOX78850.1 NEVH01019967 PNF22099.1 NNAY01001108 OXU25107.1 GDKW01000711 JAI55884.1 KZ288229 PBC31703.1 KQ414667 KOC64822.1 KK854265 PTY13929.1 KQ978473 KYM93699.1 KYM93697.1 GECL01001804 JAP04320.1 GFTR01006899 JAW09527.1 GBGD01001161 JAC87728.1 GEZM01098853 GEZM01098850 GEZM01098849 JAV53714.1 KQ980926 KYN11389.1 GL437267 EFN70727.1 LKEZ01003766 KYN50721.1 GL888102 EGI67452.1 KQ976532 KYM81551.1 ADTU01026689 GEDC01001215 JAS36083.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000008820
UP000075885
+ More
UP000076407 UP000007062 UP000075902 UP000075840 UP000075882 UP000075881 UP000075903 UP000075884 UP000075900 UP000076408 UP000075901 UP000000673 UP000075886 UP000069272 UP000075883 UP000183832 UP000075920 UP000002320 UP000095300 UP000000803 UP000002282 UP000095301 UP000007798 UP000007801 UP000268350 UP000192221 UP000037069 UP000008711 UP000001819 UP000075880 UP000008792 UP000092445 UP000001070 UP000092553 UP000091820 UP000092460 UP000092443 UP000078200 UP000092444 UP000000304 UP000001292 UP000092461 UP000008744 UP000030765 UP000027135 UP000008237 UP000076502 UP000245037 UP000053105 UP000235965 UP000215335 UP000242457 UP000005203 UP000053825 UP000078542 UP000192223 UP000078492 UP000000311 UP000078541 UP000007755 UP000078540 UP000005205
UP000076407 UP000007062 UP000075902 UP000075840 UP000075882 UP000075881 UP000075903 UP000075884 UP000075900 UP000076408 UP000075901 UP000000673 UP000075886 UP000069272 UP000075883 UP000183832 UP000075920 UP000002320 UP000095300 UP000000803 UP000002282 UP000095301 UP000007798 UP000007801 UP000268350 UP000192221 UP000037069 UP000008711 UP000001819 UP000075880 UP000008792 UP000092445 UP000001070 UP000092553 UP000091820 UP000092460 UP000092443 UP000078200 UP000092444 UP000000304 UP000001292 UP000092461 UP000008744 UP000030765 UP000027135 UP000008237 UP000076502 UP000245037 UP000053105 UP000235965 UP000215335 UP000242457 UP000005203 UP000053825 UP000078542 UP000192223 UP000078492 UP000000311 UP000078541 UP000007755 UP000078540 UP000005205
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IUV5
A0A2H1UZU7
A0A0N0PEW2
A0A194RCV6
S4PG58
A0A0L7LSA9
+ More
Q16WB3 A0A182NZP6 A0A336KX26 A0A182X5P8 Q7QHD0 A0A1S4H5T8 A0A182TPA4 A0A182HW99 A0A182KL07 A0A023EUH8 A0A182JWZ7 A0A182UVK4 A0A182N290 A0A182RW44 A0A182YB03 A0A0K8TPP4 A0A182SCF6 W8BA97 A0A0A1XHQ0 W5JN05 A0A034W0T2 A0A0K8VZ52 A0A2M4AB86 A0A182Q952 A0A182FW38 A0A182MPN7 A0A194Q7M2 A0A1Q3EWZ0 A0A1J1J3M4 A0A2M3Z269 A0A2M4BKR2 A0A182WDL1 B0WW86 A0A1I8QE28 A4V488 Q07152 B4PXZ1 T1P9Q0 B4MJ75 B3MSB7 A0A3B0JYW0 A0A1W4UNW8 A0A0L0C2R1 B3NW07 Q29IQ0 A0A182IPX8 B4MES0 A0A1A9ZRR6 B4JK62 A0A0M3QZA3 U5EIP0 A0A1A9WCQ4 A0A1B0ATC8 A0A1A9XAI2 A0A1A9UPG8 A0A1A9VPM0 A0A1A9WDG8 A0A1B0FR56 A0A1A9YA98 A0A1A9ZJM4 B4R7M3 B4IDT3 A0A1B0GGT8 B4H2U5 A0A1L8DXU5 A0A084WM01 A0A067RHP6 A0A0C9R572 E2BHJ7 A0A154P3W3 A0A2P8XQG3 A0A1Y1JWM5 A0A0M9A7J8 A0A2J7Q0K4 A0A232F3Y3 A0A0P4VV19 A0A2A3EIS3 A0A088A653 A0A0L7R1T3 A0A2R7W2W8 A0A151I6G9 A0A151I6F4 A0A0V0G8A7 A0A224XAV9 A0A069DUB9 A0A1Y1JWM0 A0A1W4WQG2 A0A151IUY9 E2A771 A0A151K3Z5 F4WE14 A0A195BAH6 A0A158NUS4 A0A1B6EDT5
Q16WB3 A0A182NZP6 A0A336KX26 A0A182X5P8 Q7QHD0 A0A1S4H5T8 A0A182TPA4 A0A182HW99 A0A182KL07 A0A023EUH8 A0A182JWZ7 A0A182UVK4 A0A182N290 A0A182RW44 A0A182YB03 A0A0K8TPP4 A0A182SCF6 W8BA97 A0A0A1XHQ0 W5JN05 A0A034W0T2 A0A0K8VZ52 A0A2M4AB86 A0A182Q952 A0A182FW38 A0A182MPN7 A0A194Q7M2 A0A1Q3EWZ0 A0A1J1J3M4 A0A2M3Z269 A0A2M4BKR2 A0A182WDL1 B0WW86 A0A1I8QE28 A4V488 Q07152 B4PXZ1 T1P9Q0 B4MJ75 B3MSB7 A0A3B0JYW0 A0A1W4UNW8 A0A0L0C2R1 B3NW07 Q29IQ0 A0A182IPX8 B4MES0 A0A1A9ZRR6 B4JK62 A0A0M3QZA3 U5EIP0 A0A1A9WCQ4 A0A1B0ATC8 A0A1A9XAI2 A0A1A9UPG8 A0A1A9VPM0 A0A1A9WDG8 A0A1B0FR56 A0A1A9YA98 A0A1A9ZJM4 B4R7M3 B4IDT3 A0A1B0GGT8 B4H2U5 A0A1L8DXU5 A0A084WM01 A0A067RHP6 A0A0C9R572 E2BHJ7 A0A154P3W3 A0A2P8XQG3 A0A1Y1JWM5 A0A0M9A7J8 A0A2J7Q0K4 A0A232F3Y3 A0A0P4VV19 A0A2A3EIS3 A0A088A653 A0A0L7R1T3 A0A2R7W2W8 A0A151I6G9 A0A151I6F4 A0A0V0G8A7 A0A224XAV9 A0A069DUB9 A0A1Y1JWM0 A0A1W4WQG2 A0A151IUY9 E2A771 A0A151K3Z5 F4WE14 A0A195BAH6 A0A158NUS4 A0A1B6EDT5
PDB
1JCN
E-value=0,
Score=1824
Ontologies
KEGG
PATHWAY
GO
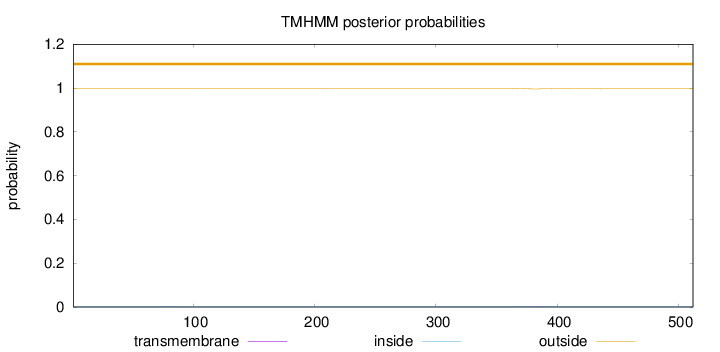
Topology
Subcellular location
Cytoplasm
Length:
512
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04899
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00220
outside
1 - 512
Population Genetic Test Statistics
Pi
3.937286
Theta
5.489165
Tajima's D
-1.09177
CLR
0.216452
CSRT
0.11734413279336
Interpretation
Uncertain
