Gene
KWMTBOMO08037 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001032
Annotation
PREDICTED:_sorting_nexin_lst-4_isoform_X1_[Papilio_xuthus]
Full name
Sorting nexin
Location in the cell
Nuclear Reliability : 2.478
Sequence
CDS
ATGGCGACCCAAGTGCAGGCTTTGTATGACTTCACTGGTGAGCCTGGCACCACAGAGATGTCCATAACTTGTGGAGAGGTGCTCACACTGCTCAACACAGAAATCGGAGAAGGCTGGTGGGAAGGAAAAAAATCTAATGGAGAAACTGGTCTATTTCCTGCTGCATATGTCAGAAAACTGACACCAGAAGAATCAATTCAAACTAAAGCTGCTCCTCCAGCACCTAGATATGATCAAGCAGCAGATGACTGGGGCGAACAACACTACAGTGCAGGAGATAACAATTACCAAAGAGGAGTTTCGCATGAAGATGGCTGGGATGATGACTGGGAAGATGATACATACTCAGAGATAGGACCAGCCCAGCAACAAAACAAGACATCGCAGTACAACCAGCAATCATCCATTGTGATTCCCGGTATGCCTATTAATGATTATAATCAACACATAGATGACACATCTTCAAATTATAGCTCATCAGTGGGCACAGTTAGAAAAAACAAATTTGCACCATCTTCAAAGATTAGTGGAGAAAGCTACCTGCTGGCAACCCTTAATGTAGAAGTACCAGACTCTGAAAAGGTCTACATTATCCAAGATGAAGATGGATATATTTGGTCCCCAATTACTCAGCCATATATTGTGACTGTGGCCTCTCCAAAAAAGGAATCTAAATTCAAAGGGATCAAGAGTTTTATAGCCTATCAACTGACTCCTTCATTCAATAATATTCAAGTGTCCAGGAGATACAAACACTTTGACTGGCTCCATGAGAGACTACAAGAAAAGTTCACACTTATTCCAATCCCCCCTCTGCCTGACAAACAGATATCTGGAAGATATGATGAGCAACTAATCGAACGCAGGCGAGTTCAGCTCCAAGAGTTTGTAGATTGGATGTGCAAGCACCCTGTGCTCTCCAGAAGCGAGGTTTGGCAACACTTCTTAACATGTACTGATGAAAAGAGATGGAAAGCTGGTAAAAGGCAAGCTGAAAAAGATAATTTGCTAGGTCTAAATTATTGTGTTTCACTAGTGGTACCTGAGAAAGCATTATTGCAATCACAAGTTGATCATATCACAGAACAGTGCCACACTTTCATTAACAGTGTCGACACAGCCATCAAGTCTGTGTCTAGCATGTGCATTGTGCAAACAAAACGTTTCCAAGGACCATACAAGAGTGACTGCCAGAAAATAGGAGAAGCATTTTATAGTCTTGGCAATGCCTTAAGCTTGGATGAAGGTACCATAGTATCAACATCTAAGCTGACTTCAGCTATTAAAATGACAGGTGGTGTCTACATTGATATTGGTAGGATGTATGATGAACAACCGAAGTATGATTGGGAGCCACTGGGTGATAAATTTCATTTATACAAAGGAATTGCAGCTAGTTTTCCTGATGTCCTGGCCAATCATAAGGGTGCAGTGCAAAAGAAAAGAGAGTGTGAAAGACTGACTGCTGAAAACAAAATGGAGGTTGCACAATTAAATGAAGTCTTGAGAAGGACCGATGTTATATCATATGCGCTTCTTGCCGAAATCAACCATTTCAAATCAGAACGGACTGTTGATATAAAAGCCACCATGCAAAAATTCTTGAAGCAACAAATCAACTTTTACAAGAAAATAGTAGATAAGTTGGAAACCACGCTAAGATACTATGATGATTAG
Protein
MATQVQALYDFTGEPGTTEMSITCGEVLTLLNTEIGEGWWEGKKSNGETGLFPAAYVRKLTPEESIQTKAAPPAPRYDQAADDWGEQHYSAGDNNYQRGVSHEDGWDDDWEDDTYSEIGPAQQQNKTSQYNQQSSIVIPGMPINDYNQHIDDTSSNYSSSVGTVRKNKFAPSSKISGESYLLATLNVEVPDSEKVYIIQDEDGYIWSPITQPYIVTVASPKKESKFKGIKSFIAYQLTPSFNNIQVSRRYKHFDWLHERLQEKFTLIPIPPLPDKQISGRYDEQLIERRRVQLQEFVDWMCKHPVLSRSEVWQHFLTCTDEKRWKAGKRQAEKDNLLGLNYCVSLVVPEKALLQSQVDHITEQCHTFINSVDTAIKSVSSMCIVQTKRFQGPYKSDCQKIGEAFYSLGNALSLDEGTIVSTSKLTSAIKMTGGVYIDIGRMYDEQPKYDWEPLGDKFHLYKGIAASFPDVLANHKGAVQKKRECERLTAENKMEVAQLNEVLRRTDVISYALLAEINHFKSERTVDIKATMQKFLKQQINFYKKIVDKLETTLRYYDD
Summary
Similarity
Belongs to the sorting nexin family.
Feature
chain Sorting nexin
Uniprot
H9IUV2
A0A194RCG0
A0A194Q7L2
A0A0L7LRJ5
A0A212FCL3
A0A2H1UZT0
+ More
A0A1E1W714 A0A2J7PXW7 A0A2J7PXU3 A0A067R133 A0A1B0CA89 A0A1B0D563 A0A1L8DVN0 U5EZN9 A0A1B6HWD0 A0A1Y1MFI1 A0A182RR17 A0A1Q3F3T1 A0A182PUK8 A0A1Q3F4K7 A0A1Q3F504 A0A1W4X5B2 A0A3B0K193 A0A182YL26 A0A182HH61 A0A182L4N4 A0A182N3L0 A0A182X6Q6 Q29EF9 B4H1Y9 A0A1B6DKN3 A0A182TXV3 A0A182UTP8 A0A182FVT9 Q7PZ64 A0A2M4ACI2 A0A2M4AC76 A0A182W2E0 W5J6I3 A0A2M4AC81 A0A084WJ38 E2BN35 A0A0A1XA87 A0A023EU45 A0A182GJ82 D3TRI4 W8BRJ0 A0A034VDN6 A0A1A9XHI9 A0A1B0BHV3 A0A2M3ZHD8 A0A182MA38 B0WRZ3 A0A336LU90 A0A0K8W2G7 A0A182IRN7 B3NGK5 B4HL95 A0A0J9RSV4 Q9NCC3 B4PED5 A0A1A9W7H7 A0A2M4BIC8 A0A2M4BIB4 B4QNM9 A0A1B6M8Q9 B4LBR4 A0A0M5JB09 A0A1W4UKY9 B4L998 A0A2M3Z5V5 A0A026WNS9 A0A1I8MXY7 A0A158NQY9 A0A1B6GZJ7 B4MX98 A0A195B0E8 A0A310SNI3 E0V9Z0 A0A151WMD4 A0A0P6E5K5 A0A0P5JN71 A0A0P5SAN3 A0A0P6C2I7 A0A0P5PU23 A0A0P5H6L7 A0A0P5MN77 A0A0P5QNC6 A0A0L7R3B0 A0A0P5MYJ2 A0A0P5RTG8 A0A0P5RDL1 A0A1B6HV50 B4J1I5 A0A0P5N939 B3MA80 E9H8V4 A0A0P5WAU5 A0A1A9Z4L2
A0A1E1W714 A0A2J7PXW7 A0A2J7PXU3 A0A067R133 A0A1B0CA89 A0A1B0D563 A0A1L8DVN0 U5EZN9 A0A1B6HWD0 A0A1Y1MFI1 A0A182RR17 A0A1Q3F3T1 A0A182PUK8 A0A1Q3F4K7 A0A1Q3F504 A0A1W4X5B2 A0A3B0K193 A0A182YL26 A0A182HH61 A0A182L4N4 A0A182N3L0 A0A182X6Q6 Q29EF9 B4H1Y9 A0A1B6DKN3 A0A182TXV3 A0A182UTP8 A0A182FVT9 Q7PZ64 A0A2M4ACI2 A0A2M4AC76 A0A182W2E0 W5J6I3 A0A2M4AC81 A0A084WJ38 E2BN35 A0A0A1XA87 A0A023EU45 A0A182GJ82 D3TRI4 W8BRJ0 A0A034VDN6 A0A1A9XHI9 A0A1B0BHV3 A0A2M3ZHD8 A0A182MA38 B0WRZ3 A0A336LU90 A0A0K8W2G7 A0A182IRN7 B3NGK5 B4HL95 A0A0J9RSV4 Q9NCC3 B4PED5 A0A1A9W7H7 A0A2M4BIC8 A0A2M4BIB4 B4QNM9 A0A1B6M8Q9 B4LBR4 A0A0M5JB09 A0A1W4UKY9 B4L998 A0A2M3Z5V5 A0A026WNS9 A0A1I8MXY7 A0A158NQY9 A0A1B6GZJ7 B4MX98 A0A195B0E8 A0A310SNI3 E0V9Z0 A0A151WMD4 A0A0P6E5K5 A0A0P5JN71 A0A0P5SAN3 A0A0P6C2I7 A0A0P5PU23 A0A0P5H6L7 A0A0P5MN77 A0A0P5QNC6 A0A0L7R3B0 A0A0P5MYJ2 A0A0P5RTG8 A0A0P5RDL1 A0A1B6HV50 B4J1I5 A0A0P5N939 B3MA80 E9H8V4 A0A0P5WAU5 A0A1A9Z4L2
Pubmed
19121390
26354079
26227816
22118469
24845553
28004739
+ More
25244985 20966253 15632085 17994087 12364791 20920257 23761445 24438588 20798317 25830018 24945155 26483478 20353571 24495485 25348373 22936249 10823906 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24508170 30249741 25315136 21347285 20566863 21292972
25244985 20966253 15632085 17994087 12364791 20920257 23761445 24438588 20798317 25830018 24945155 26483478 20353571 24495485 25348373 22936249 10823906 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24508170 30249741 25315136 21347285 20566863 21292972
EMBL
BABH01000428
KQ460397
KPJ15287.1
KQ459439
KPJ00985.1
JTDY01000244
+ More
KOB78080.1 AGBW02009165 OWR51485.1 ODYU01000029 SOQ34107.1 GDQN01009049 GDQN01008274 JAT82005.1 JAT82780.1 NEVH01020852 PNF21154.1 PNF21155.1 KK852819 KDR15627.1 AJWK01003422 AJVK01003327 GFDF01003596 JAV10488.1 GANO01000436 JAB59435.1 GECU01028725 JAS78981.1 GEZM01037444 JAV82067.1 GFDL01012834 JAV22211.1 GFDL01012555 JAV22490.1 GFDL01012403 JAV22642.1 OUUW01000002 SPP77158.1 APCN01002389 CH379070 EAL30102.1 CH479203 EDW30341.1 GEDC01011045 JAS26253.1 AAAB01008986 EAA00038.4 GGFK01005111 MBW38432.1 GGFK01005074 MBW38395.1 ADMH02001988 ETN60067.1 GGFK01005083 MBW38404.1 ATLV01023972 KE525347 KFB50232.1 GL449382 EFN82939.1 GBXI01006704 JAD07588.1 JXUM01072950 GAPW01000690 KQ562749 JAC12908.1 KXJ75217.1 JXUM01067638 KQ562460 KXJ75862.1 CCAG010006049 EZ424036 ADD20312.1 GAMC01010674 JAB95881.1 GAKP01019052 JAC39900.1 JXJN01014603 GGFM01007178 MBW27929.1 AXCM01010654 DS232063 EDS33611.1 UFQT01000114 SSX20229.1 GDHF01006941 JAI45373.1 CH954178 EDV51241.1 CH480815 EDW40914.1 CM002912 KMY98737.1 AF223381 AE014296 AY069705 AAF34701.1 AAF50219.2 AAL39850.1 CM000159 EDW92973.1 GGFJ01003656 MBW52797.1 GGFJ01003655 MBW52796.1 CM000363 EDX09896.1 GEBQ01007679 JAT32298.1 CH940647 EDW68691.1 CP012525 ALC43731.1 CH933816 EDW17273.1 GGFM01003151 MBW23902.1 KK107139 QOIP01000003 EZA57712.1 RLU24345.1 ADTU01023705 GECZ01001923 JAS67846.1 CH963876 EDW76931.1 KQ976691 KYM77953.1 KQ760801 OAD58972.1 DS235001 EEB10196.1 KQ982944 KYQ48978.1 GDIQ01092089 GDIQ01079499 LRGB01001005 JAN15238.1 KZS14197.1 GDIQ01196151 JAK55574.1 GDIQ01096242 JAL55484.1 GDIP01022116 JAM81599.1 GDIQ01124182 JAL27544.1 GDIQ01232138 JAK19587.1 GDIQ01161072 JAK90653.1 GDIQ01121727 JAL29999.1 KQ414663 KOC65266.1 GDIQ01157671 JAK94054.1 GDIQ01105645 JAL46081.1 GDIQ01121726 JAL30000.1 GECU01029149 JAS78557.1 CH916366 EDV95876.1 GDIQ01152624 JAK99101.1 CH902618 EDV39094.1 GL732606 EFX71786.1 GDIQ01140889 GDIP01088709 GDIP01083598 JAL10837.1 JAM15006.1
KOB78080.1 AGBW02009165 OWR51485.1 ODYU01000029 SOQ34107.1 GDQN01009049 GDQN01008274 JAT82005.1 JAT82780.1 NEVH01020852 PNF21154.1 PNF21155.1 KK852819 KDR15627.1 AJWK01003422 AJVK01003327 GFDF01003596 JAV10488.1 GANO01000436 JAB59435.1 GECU01028725 JAS78981.1 GEZM01037444 JAV82067.1 GFDL01012834 JAV22211.1 GFDL01012555 JAV22490.1 GFDL01012403 JAV22642.1 OUUW01000002 SPP77158.1 APCN01002389 CH379070 EAL30102.1 CH479203 EDW30341.1 GEDC01011045 JAS26253.1 AAAB01008986 EAA00038.4 GGFK01005111 MBW38432.1 GGFK01005074 MBW38395.1 ADMH02001988 ETN60067.1 GGFK01005083 MBW38404.1 ATLV01023972 KE525347 KFB50232.1 GL449382 EFN82939.1 GBXI01006704 JAD07588.1 JXUM01072950 GAPW01000690 KQ562749 JAC12908.1 KXJ75217.1 JXUM01067638 KQ562460 KXJ75862.1 CCAG010006049 EZ424036 ADD20312.1 GAMC01010674 JAB95881.1 GAKP01019052 JAC39900.1 JXJN01014603 GGFM01007178 MBW27929.1 AXCM01010654 DS232063 EDS33611.1 UFQT01000114 SSX20229.1 GDHF01006941 JAI45373.1 CH954178 EDV51241.1 CH480815 EDW40914.1 CM002912 KMY98737.1 AF223381 AE014296 AY069705 AAF34701.1 AAF50219.2 AAL39850.1 CM000159 EDW92973.1 GGFJ01003656 MBW52797.1 GGFJ01003655 MBW52796.1 CM000363 EDX09896.1 GEBQ01007679 JAT32298.1 CH940647 EDW68691.1 CP012525 ALC43731.1 CH933816 EDW17273.1 GGFM01003151 MBW23902.1 KK107139 QOIP01000003 EZA57712.1 RLU24345.1 ADTU01023705 GECZ01001923 JAS67846.1 CH963876 EDW76931.1 KQ976691 KYM77953.1 KQ760801 OAD58972.1 DS235001 EEB10196.1 KQ982944 KYQ48978.1 GDIQ01092089 GDIQ01079499 LRGB01001005 JAN15238.1 KZS14197.1 GDIQ01196151 JAK55574.1 GDIQ01096242 JAL55484.1 GDIP01022116 JAM81599.1 GDIQ01124182 JAL27544.1 GDIQ01232138 JAK19587.1 GDIQ01161072 JAK90653.1 GDIQ01121727 JAL29999.1 KQ414663 KOC65266.1 GDIQ01157671 JAK94054.1 GDIQ01105645 JAL46081.1 GDIQ01121726 JAL30000.1 GECU01029149 JAS78557.1 CH916366 EDV95876.1 GDIQ01152624 JAK99101.1 CH902618 EDV39094.1 GL732606 EFX71786.1 GDIQ01140889 GDIP01088709 GDIP01083598 JAL10837.1 JAM15006.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000007151
UP000235965
+ More
UP000027135 UP000092461 UP000092462 UP000075900 UP000075885 UP000192223 UP000268350 UP000076408 UP000075840 UP000075882 UP000075884 UP000076407 UP000001819 UP000008744 UP000075902 UP000075903 UP000069272 UP000007062 UP000075920 UP000000673 UP000030765 UP000008237 UP000069940 UP000249989 UP000092444 UP000092443 UP000092460 UP000075883 UP000002320 UP000075880 UP000008711 UP000001292 UP000000803 UP000002282 UP000091820 UP000000304 UP000008792 UP000092553 UP000192221 UP000009192 UP000053097 UP000279307 UP000095301 UP000005205 UP000007798 UP000078540 UP000009046 UP000075809 UP000076858 UP000053825 UP000001070 UP000007801 UP000000305 UP000092445
UP000027135 UP000092461 UP000092462 UP000075900 UP000075885 UP000192223 UP000268350 UP000076408 UP000075840 UP000075882 UP000075884 UP000076407 UP000001819 UP000008744 UP000075902 UP000075903 UP000069272 UP000007062 UP000075920 UP000000673 UP000030765 UP000008237 UP000069940 UP000249989 UP000092444 UP000092443 UP000092460 UP000075883 UP000002320 UP000075880 UP000008711 UP000001292 UP000000803 UP000002282 UP000091820 UP000000304 UP000008792 UP000092553 UP000192221 UP000009192 UP000053097 UP000279307 UP000095301 UP000005205 UP000007798 UP000078540 UP000009046 UP000075809 UP000076858 UP000053825 UP000001070 UP000007801 UP000000305 UP000092445
Interpro
Gene 3D
ProteinModelPortal
H9IUV2
A0A194RCG0
A0A194Q7L2
A0A0L7LRJ5
A0A212FCL3
A0A2H1UZT0
+ More
A0A1E1W714 A0A2J7PXW7 A0A2J7PXU3 A0A067R133 A0A1B0CA89 A0A1B0D563 A0A1L8DVN0 U5EZN9 A0A1B6HWD0 A0A1Y1MFI1 A0A182RR17 A0A1Q3F3T1 A0A182PUK8 A0A1Q3F4K7 A0A1Q3F504 A0A1W4X5B2 A0A3B0K193 A0A182YL26 A0A182HH61 A0A182L4N4 A0A182N3L0 A0A182X6Q6 Q29EF9 B4H1Y9 A0A1B6DKN3 A0A182TXV3 A0A182UTP8 A0A182FVT9 Q7PZ64 A0A2M4ACI2 A0A2M4AC76 A0A182W2E0 W5J6I3 A0A2M4AC81 A0A084WJ38 E2BN35 A0A0A1XA87 A0A023EU45 A0A182GJ82 D3TRI4 W8BRJ0 A0A034VDN6 A0A1A9XHI9 A0A1B0BHV3 A0A2M3ZHD8 A0A182MA38 B0WRZ3 A0A336LU90 A0A0K8W2G7 A0A182IRN7 B3NGK5 B4HL95 A0A0J9RSV4 Q9NCC3 B4PED5 A0A1A9W7H7 A0A2M4BIC8 A0A2M4BIB4 B4QNM9 A0A1B6M8Q9 B4LBR4 A0A0M5JB09 A0A1W4UKY9 B4L998 A0A2M3Z5V5 A0A026WNS9 A0A1I8MXY7 A0A158NQY9 A0A1B6GZJ7 B4MX98 A0A195B0E8 A0A310SNI3 E0V9Z0 A0A151WMD4 A0A0P6E5K5 A0A0P5JN71 A0A0P5SAN3 A0A0P6C2I7 A0A0P5PU23 A0A0P5H6L7 A0A0P5MN77 A0A0P5QNC6 A0A0L7R3B0 A0A0P5MYJ2 A0A0P5RTG8 A0A0P5RDL1 A0A1B6HV50 B4J1I5 A0A0P5N939 B3MA80 E9H8V4 A0A0P5WAU5 A0A1A9Z4L2
A0A1E1W714 A0A2J7PXW7 A0A2J7PXU3 A0A067R133 A0A1B0CA89 A0A1B0D563 A0A1L8DVN0 U5EZN9 A0A1B6HWD0 A0A1Y1MFI1 A0A182RR17 A0A1Q3F3T1 A0A182PUK8 A0A1Q3F4K7 A0A1Q3F504 A0A1W4X5B2 A0A3B0K193 A0A182YL26 A0A182HH61 A0A182L4N4 A0A182N3L0 A0A182X6Q6 Q29EF9 B4H1Y9 A0A1B6DKN3 A0A182TXV3 A0A182UTP8 A0A182FVT9 Q7PZ64 A0A2M4ACI2 A0A2M4AC76 A0A182W2E0 W5J6I3 A0A2M4AC81 A0A084WJ38 E2BN35 A0A0A1XA87 A0A023EU45 A0A182GJ82 D3TRI4 W8BRJ0 A0A034VDN6 A0A1A9XHI9 A0A1B0BHV3 A0A2M3ZHD8 A0A182MA38 B0WRZ3 A0A336LU90 A0A0K8W2G7 A0A182IRN7 B3NGK5 B4HL95 A0A0J9RSV4 Q9NCC3 B4PED5 A0A1A9W7H7 A0A2M4BIC8 A0A2M4BIB4 B4QNM9 A0A1B6M8Q9 B4LBR4 A0A0M5JB09 A0A1W4UKY9 B4L998 A0A2M3Z5V5 A0A026WNS9 A0A1I8MXY7 A0A158NQY9 A0A1B6GZJ7 B4MX98 A0A195B0E8 A0A310SNI3 E0V9Z0 A0A151WMD4 A0A0P6E5K5 A0A0P5JN71 A0A0P5SAN3 A0A0P6C2I7 A0A0P5PU23 A0A0P5H6L7 A0A0P5MN77 A0A0P5QNC6 A0A0L7R3B0 A0A0P5MYJ2 A0A0P5RTG8 A0A0P5RDL1 A0A1B6HV50 B4J1I5 A0A0P5N939 B3MA80 E9H8V4 A0A0P5WAU5 A0A1A9Z4L2
PDB
4AKV
E-value=2.89425e-77,
Score=736
Ontologies
GO
GO:0035091
GO:0000278
GO:0015031
GO:0000045
GO:0005938
GO:0030032
GO:0001956
GO:0043025
GO:0048471
GO:0071212
GO:0042734
GO:1990709
GO:0019900
GO:0071456
GO:0006886
GO:0000281
GO:0006897
GO:0005623
GO:0005546
GO:0005886
GO:0043195
GO:0005515
GO:0005975
GO:0016021
GO:0016020
GO:0005524
GO:0006412
GO:0016876
GO:0043039
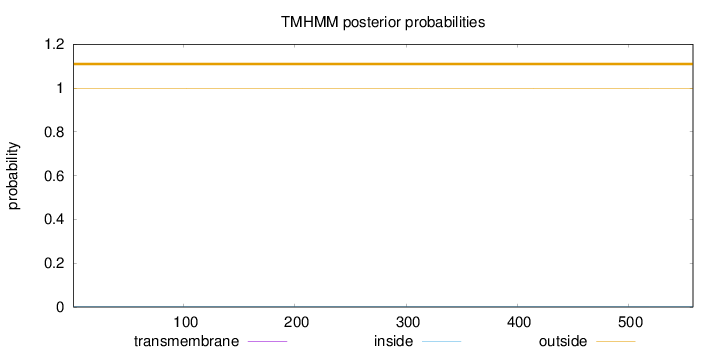
Topology
Length:
558
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00213
Exp number, first 60 AAs:
0.00049
Total prob of N-in:
0.00234
outside
1 - 558
Population Genetic Test Statistics
Pi
13.444147
Theta
11.077705
Tajima's D
-0.193951
CLR
1.338827
CSRT
0.326033698315084
Interpretation
Uncertain
