Gene
KWMTBOMO08033
Pre Gene Modal
BGIBMGA001028
Annotation
PREDICTED:_tetratricopeptide_repeat_protein_17_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.218
Sequence
CDS
ATGAACTACATTAACTTTTTATGGTTATTCGGTTTGTGTACCGTTCATGCAACGACGATTGCCACACACTGGATGGTTACTGAAACAGGACTGATACAACCTCGAGCTGACTCTCCATTCATAATGGAACATCCCAATGATCTATTAGCATTCTTAAATCAAGACACGAGATGGGATAACATCCACAACCTGTATTATGATTTATATAACCGCCAACAAATCATTGATCAGCTTTGGCTTGACATTGAAAAGGGTACTGAAGTGACAACTATACTCACTTCCAATCAACACTGTGTTAGAGTGGGACAGCTGAATATTATAGATTGGTATACATCATTCTTAGAAGATGGCAAATCGAAAGGAGTACCCGATGAGGATTTTTTACTTAGAGAGACTGATTATGGAGATACCGATGTACCTGATTGTAAAAAGATATCTTCATTGACATTTAGCATGTTTGCCTTTGAGCATTTAGAGGGTTTGAATCAAAGAGAGAATTTATCAATTGATCCTGAACTGTCACTCCCAGAACAAATAACTCCCATTATGACTGTTGATCCATTTGGTCACTGGCTTGCAGCAATGCTAAGGCGGAATAACTCTTCCTGGCTTCATTATAACATGGCTTCTCTATATTGGAGAGTAAAAGGAAATGCACCTAAAGCTATGGAGTGCAGCAGAAGAGCTGTTCATTACGCTCCTAGAAAGTATAAAGACATAGCTTTATTGAGCATGGGTACAATACTTCACCGCAGCAAAAGAACCGAAGATGCTATAGTAGTTCTCAGAGCAGCTACTGATCACGATCCGACATTCACATACAATCATTTCTCTCTAGCCAATGCCTATGCTGTGTTCGGTGACTTCAATAGTTCTTTAAAACATTTCGCTGAGTGTCTCAAACTGAATCCTACTTTTGAATTGGCAGTGCGACATAAAATTGCAGTTTTATGTCACCTTTATGTGTCTGATACAATTGAAGAGTTAAAGGAAACCCTACAAAAACTGCGCCAAGAGCTAATAGAATATGCAAGAAGAGAGAGTGAATGGCTAAAAAGTCAAGCTGCGTTTTTGAGGACAATGAAACATGAAGAAGAATTTGATTACCGAAATATAGACAATAATTGTAAAAAGATGACCAATATAACTGGTTTAAAAATAAAAGAACTGAAACTACAAGGAGACAGAAACTCATTGCTCCAATATTTCTTGGACAGTCCGATGTACAACGATCAATGGTTGAAGGAGAAAGGTGTACTCGCAATAGAATCGGCCGGGAATCTCCAGAAACTGGTCAAACACATCCGGCAGAATTCTCCGAAGGAAGCAACACTGAAAATCCGACCAGAGAAACTGAAAGTAGCTGAAGTGGAAGTATTAAAAGAGATTGGTCCAACACCAATATTCCCTGATATTTATACAGATATGAAAGAAAAAACATTGATTGAAGTTTTGGCTGCGGCAGAATTTCGAAAGAACAACGAAAGAAAACCAGGTCAGAATAAAAAAGAAAGTGAAACTTCTCCATTCGAAACCGGTGTAATGATGTATCCGCCTACAATAAAAATCAATAGGAATCATCAAGATTTCGACGTCGACGCTGATTGGCCCTCAACGGATTTTTGTAAAGAGAATTCTCCAAACTTCCCCGAGAATCTTGAGGCTATATATCCGACGTTCATGCCCTTTGAAAATAAAGGAATCAGGCTGATGGCTCTATTGACGGATATGATAGGAGTTCCGGCCTCCGTCGAGCATGAGCTGCCTTGGCACCCCCCCACCTGCCCGCCGGATAAGGATATCACACTATTCACACAAAAGAAAGTCAAGCCTCAGTTCTCGAGCGAGGTGGCCGGCGTGGGCCCGCTGCGCCGCAAGCTGCACGAGTACGTCGCGGACGGAGACCTCGCCGCCGCGCAGCGCATGCAGGACGCGGAGATCGGACAGAGGATATACGCGGCCATGCAGAAGAAACTAGCGCCGAAATGGATCCTGTACACGTTGTCGTCGCTGTACTGGCGCGTGCGAAGCAACAATATAAACGCTTTACACTGCCTTCTCAACGCCGGCAGGACTGTCGAAAATAAATACAGAGATATAGTGTTGACTTCAGTAGCGTCGATATATCTGGAGATGGGGTACTTCGACGAGGCCTTCACCGTTGCCGAAGAAGCGTTCAGACTGAGTCTGTTTGAGCCAGCAACAAACTTTATACTGGCAGAACTGAATATGATTAATAAACATCGGAAGACGCATTTGATTCACTTGAAGCAAGTTTTAAGAGTAGAACCCTCTTTTATGGGAGGAATCGTTCGGAAACTTTTGATCGGATGGGCTTGTATACTAAAGCAAGTTAACACTGTGAACGAAGTGGAACTCGAGGAGGGTGAGGTTTGTTCTCGGGTCGAGACGGGAACGAACATGGTCTGCGAAAAGGACGGCTCCAACTGTCAAATGTCCAACGTTAAATGCTTTAGTAGCCGTGAACGCGAGAGTGGGGGCGGCGCTATGGTTAGAATGTTAGAGCTAAAAGACGAAAACTCACAACGACCCGATGTCGAACGCATAGAGTCGGATGTGTTCGATCACGTGATAAGGAACATGCCCGCCGAGTCCTCCGATAAGGTGGCCCATTTAAAAAACTACGACGCCATGCTGAGGACCATGGACAAATCATTGAACGTCTGCGGGAAGGATGGATGTTATTCTGAAATCCAACTCGACAATATATTATTAGAAGAAGAGGACTGTTCAAATCAACATTTGCAATTGAGCTATTGGTTGCACGTCAGTTTTAGACAGATGCTAAGCGAAGCCGATCTCGGGCTCGTGCAAGAGATAACAGCGACTCCGCCGGCCGGAAGAAAAGTTCCCGATTGCAGAGCGCCCACTGTACAGGACTTCTTCATCGACCGCATGATAACCGTAGACACGGCCGGTTGGGAACCGGTGTTCAGTCTAATGCACCAGCTCGCCGAGATGTTCAATTTCTACGATTATACGACGCTCGGATCCAAAATCGCCAAATACGTTGAATCTCACCCGAGCTCGTGGTCGGCATTGATGTGGGCGGGGTGGTGGTGCGGGGCGGGGGGCCGCGGGCCGTGCGCCGCGCGCTGCCTGTGCGCCGCGCATGCGCACGCGCCCGCGCACGCACACACGTACGCGCTCTCCGGGCTCGTCGCTCTGCTGCACATGCAATCGAAACAAAAGGACGCTAAAGATATATCGTATCTGGCTTTGTACTCTTCGCCTAAAAGTAAAAATGAAGCGTTTTTGGTGGCCTTGTCTCATACTTTACTGTCGAAGACAGCCTTTTTCATTTGA
Protein
MNYINFLWLFGLCTVHATTIATHWMVTETGLIQPRADSPFIMEHPNDLLAFLNQDTRWDNIHNLYYDLYNRQQIIDQLWLDIEKGTEVTTILTSNQHCVRVGQLNIIDWYTSFLEDGKSKGVPDEDFLLRETDYGDTDVPDCKKISSLTFSMFAFEHLEGLNQRENLSIDPELSLPEQITPIMTVDPFGHWLAAMLRRNNSSWLHYNMASLYWRVKGNAPKAMECSRRAVHYAPRKYKDIALLSMGTILHRSKRTEDAIVVLRAATDHDPTFTYNHFSLANAYAVFGDFNSSLKHFAECLKLNPTFELAVRHKIAVLCHLYVSDTIEELKETLQKLRQELIEYARRESEWLKSQAAFLRTMKHEEEFDYRNIDNNCKKMTNITGLKIKELKLQGDRNSLLQYFLDSPMYNDQWLKEKGVLAIESAGNLQKLVKHIRQNSPKEATLKIRPEKLKVAEVEVLKEIGPTPIFPDIYTDMKEKTLIEVLAAAEFRKNNERKPGQNKKESETSPFETGVMMYPPTIKINRNHQDFDVDADWPSTDFCKENSPNFPENLEAIYPTFMPFENKGIRLMALLTDMIGVPASVEHELPWHPPTCPPDKDITLFTQKKVKPQFSSEVAGVGPLRRKLHEYVADGDLAAAQRMQDAEIGQRIYAAMQKKLAPKWILYTLSSLYWRVRSNNINALHCLLNAGRTVENKYRDIVLTSVASIYLEMGYFDEAFTVAEEAFRLSLFEPATNFILAELNMINKHRKTHLIHLKQVLRVEPSFMGGIVRKLLIGWACILKQVNTVNEVELEEGEVCSRVETGTNMVCEKDGSNCQMSNVKCFSSRERESGGGAMVRMLELKDENSQRPDVERIESDVFDHVIRNMPAESSDKVAHLKNYDAMLRTMDKSLNVCGKDGCYSEIQLDNILLEEEDCSNQHLQLSYWLHVSFRQMLSEADLGLVQEITATPPAGRKVPDCRAPTVQDFFIDRMITVDTAGWEPVFSLMHQLAEMFNFYDYTTLGSKIAKYVESHPSSWSALMWAGWWCGAGGRGPCAARCLCAAHAHAPAHAHTYALSGLVALLHMQSKQKDAKDISYLALYSSPKSKNEAFLVALSHTLLSKTAFFI
Summary
Uniprot
A0A2H1WMS3
A0A194Q7K7
A0A194RCF5
A0A212FN99
A0A1Y1NDP2
A0A1W4XEL0
+ More
A0A1W4X517 A0A067RM71 A0A2J7R942 A0A0T6BE90 D2A0N1 A0A232ERL5 A0A3L8DVW5 A0A3S1B591 A0A026W3Q5 E2AZ97 A0A195DRF0 A0A158NBF6 A0A195FCT0 A0A1A8P660 A0A1A8LEQ9 A0A1A8PXW8 A0A1A8NLT6 A0A1A8JL94 V4A941 A0A1A7YCY8 A0A1A8HGC3 A0A158RD00 A0A1I9G3H5 A0A1I9G3J8 A0A0K0IU32 A0A1I9G3P3
A0A1W4X517 A0A067RM71 A0A2J7R942 A0A0T6BE90 D2A0N1 A0A232ERL5 A0A3L8DVW5 A0A3S1B591 A0A026W3Q5 E2AZ97 A0A195DRF0 A0A158NBF6 A0A195FCT0 A0A1A8P660 A0A1A8LEQ9 A0A1A8PXW8 A0A1A8NLT6 A0A1A8JL94 V4A941 A0A1A7YCY8 A0A1A8HGC3 A0A158RD00 A0A1I9G3H5 A0A1I9G3J8 A0A0K0IU32 A0A1I9G3P3
Pubmed
EMBL
ODYU01009740
SOQ54369.1
KQ459439
KPJ00980.1
KQ460397
KPJ15282.1
+ More
AGBW02006772 OWR55193.1 GEZM01005645 JAV96013.1 KK852543 KDR21670.1 NEVH01006721 PNF37345.1 LJIG01001425 KRT85515.1 KQ971338 EFA02534.2 NNAY01002580 OXU21005.1 QOIP01000004 RLU24109.1 RQTK01000404 RUS80218.1 KK107453 EZA50628.1 GL444149 EFN61240.1 KQ980581 KYN15443.1 ADTU01011036 KQ981673 KYN38196.1 HAEG01006468 SBR76810.1 HAEF01005705 SBR43087.1 HAEH01009029 SBR86086.1 HAEI01000027 SBR69859.1 HAED01013178 HAEE01000831 SBR20847.1 KB202237 ESO91575.1 HADX01006150 SBP28382.1 HAEC01014047 SBQ82264.1 UYYF01004820 VDN07175.1 LN856993 CDP98255.1 CDP98253.1 CDP98254.1
AGBW02006772 OWR55193.1 GEZM01005645 JAV96013.1 KK852543 KDR21670.1 NEVH01006721 PNF37345.1 LJIG01001425 KRT85515.1 KQ971338 EFA02534.2 NNAY01002580 OXU21005.1 QOIP01000004 RLU24109.1 RQTK01000404 RUS80218.1 KK107453 EZA50628.1 GL444149 EFN61240.1 KQ980581 KYN15443.1 ADTU01011036 KQ981673 KYN38196.1 HAEG01006468 SBR76810.1 HAEF01005705 SBR43087.1 HAEH01009029 SBR86086.1 HAEI01000027 SBR69859.1 HAED01013178 HAEE01000831 SBR20847.1 KB202237 ESO91575.1 HADX01006150 SBP28382.1 HAEC01014047 SBQ82264.1 UYYF01004820 VDN07175.1 LN856993 CDP98255.1 CDP98253.1 CDP98254.1
Proteomes
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2H1WMS3
A0A194Q7K7
A0A194RCF5
A0A212FN99
A0A1Y1NDP2
A0A1W4XEL0
+ More
A0A1W4X517 A0A067RM71 A0A2J7R942 A0A0T6BE90 D2A0N1 A0A232ERL5 A0A3L8DVW5 A0A3S1B591 A0A026W3Q5 E2AZ97 A0A195DRF0 A0A158NBF6 A0A195FCT0 A0A1A8P660 A0A1A8LEQ9 A0A1A8PXW8 A0A1A8NLT6 A0A1A8JL94 V4A941 A0A1A7YCY8 A0A1A8HGC3 A0A158RD00 A0A1I9G3H5 A0A1I9G3J8 A0A0K0IU32 A0A1I9G3P3
A0A1W4X517 A0A067RM71 A0A2J7R942 A0A0T6BE90 D2A0N1 A0A232ERL5 A0A3L8DVW5 A0A3S1B591 A0A026W3Q5 E2AZ97 A0A195DRF0 A0A158NBF6 A0A195FCT0 A0A1A8P660 A0A1A8LEQ9 A0A1A8PXW8 A0A1A8NLT6 A0A1A8JL94 V4A941 A0A1A7YCY8 A0A1A8HGC3 A0A158RD00 A0A1I9G3H5 A0A1I9G3J8 A0A0K0IU32 A0A1I9G3P3
PDB
2HYZ
E-value=0.000212429,
Score=110
Ontologies
GO
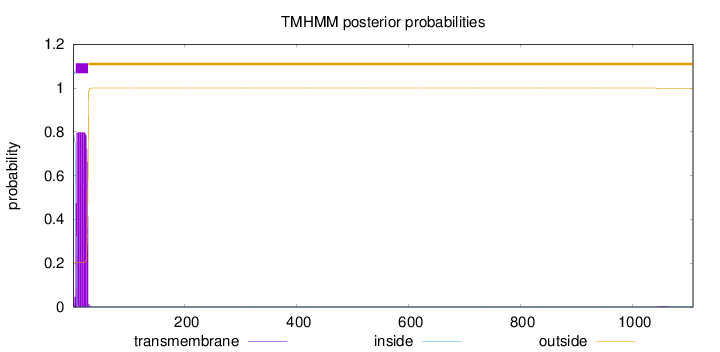
Topology
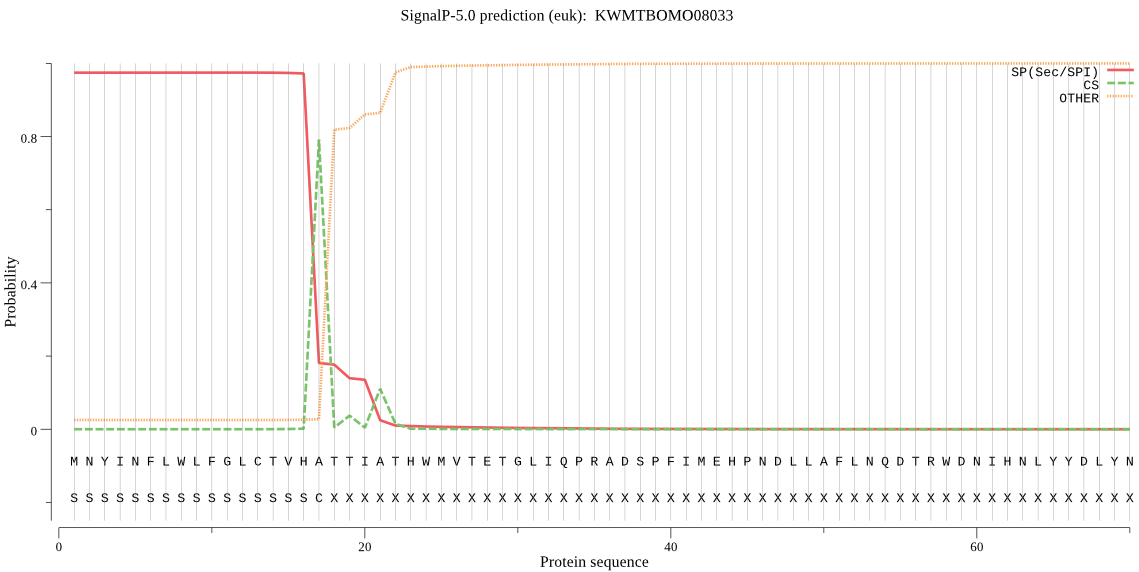
SignalP
Position: 1 - 17,
Likelihood: 0.974298
Length:
1108
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.2879299999999
Exp number, first 60 AAs:
17.21455
Total prob of N-in:
0.79770
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 1108
Population Genetic Test Statistics
Pi
17.94653
Theta
17.334261
Tajima's D
-0.771709
CLR
1.414547
CSRT
0.181340932953352
Interpretation
Uncertain
